Expand Your Single-Cell Genomics Studies with Proteomics
Single-cell genomics technologies enable researchers to study the transcription profiles of individual cells. They offer profound insights into cell-to-cell variability but are especially powerful when combined with simultaneous proteomic analysis. Here, we explain how DNA-barcoded antibodies can be used to couple specific detection of protein markers with transcriptome profiling and comment on how this approach adds value to scientific research. We also note the critical importance of robust software to multiomics workflows.
Single-cell analysis provides unique insights
Before the advent of DNA-barcoded antibodies such as BioLegend’s TotalSeq™, the analysis of single cells was broadly divided into two categories based on the type of target being studied. Single-cell protein analysis relied on fluorescently labeled antibodies to evaluate the expression of cell surface and intracellular proteins by flow cytometry,while single-cell genomics technologies such as single-cell RNA sequencing (scRNA-seq) have been widely used to quantify mRNA.
Although both approaches can identify novel cell types and reveal events that might be overlooked when cells are pooled for analysis, they each have inherent limitations. Issues relating to spectral overlap restrict the number of targets that can reliably be analyzed in parallel by flow cytometry. And it can be challenging to differentiate cells that are transcriptomically similar using techniques such as scRNA-seq.
Multiomics approaches enhance understanding
Multiomics studies combining several datasets have long been recognized for their potential to enhance researchers’ understanding of a diversity of biological processes. Yet being able to simultaneously perform proteomics and transcriptomics in single cells promises even deeper insights. For example, quantifying surface protein expression and correlating this to cellular mRNA levels enables clearer definition of the immune cells involved in response to infection.
Until recently, interrogating single cells in this way was impossible. But with the development of a new multiomics technique known as Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq), this is no longer the case.
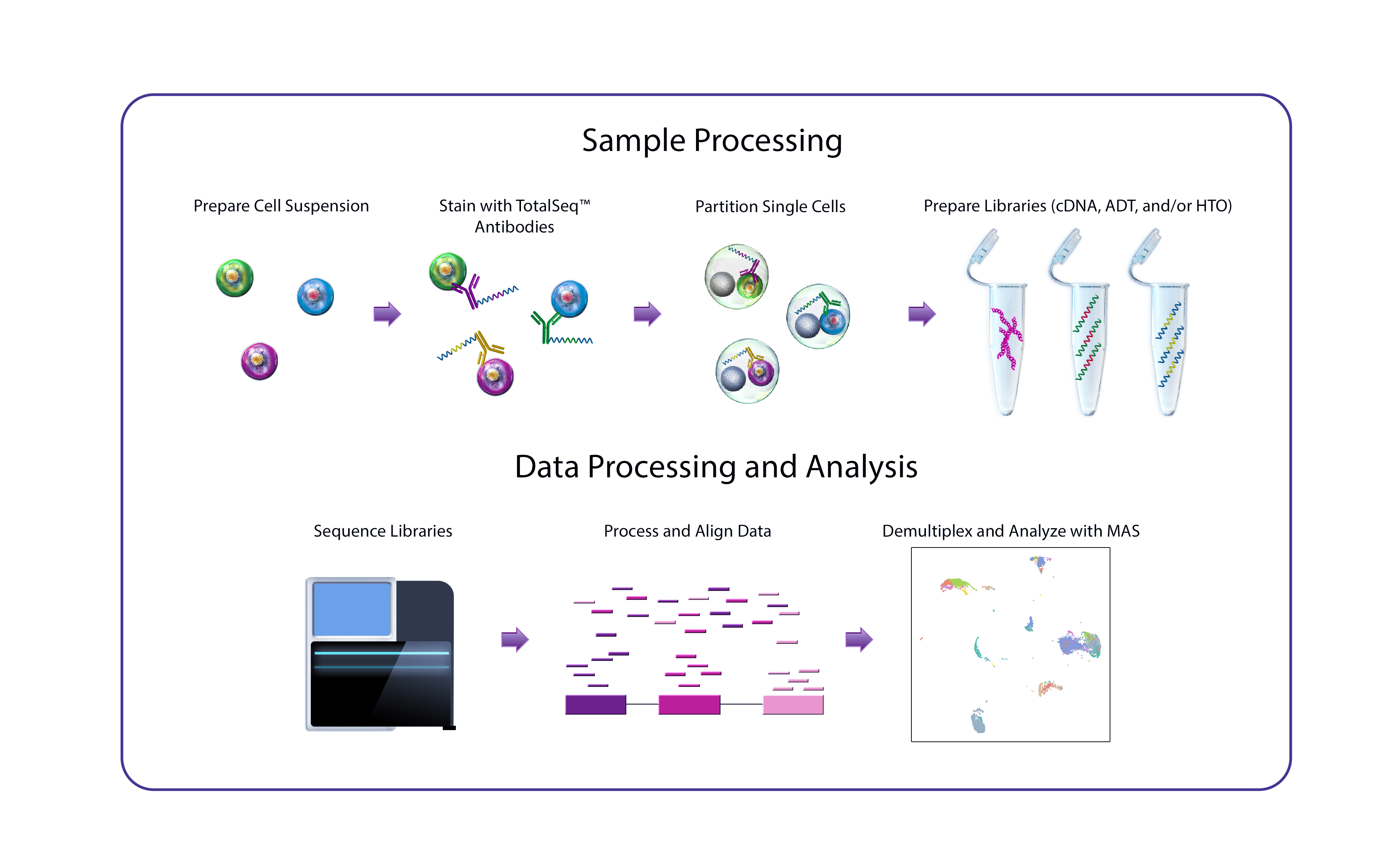
DNA-barcoded antibodies make single-cell multiomics a reality
CITE-seq, developed at the New York Genome Center’s (NYGC) Technology Innovation Lab, uses DNA-barcoded antibodies to convert protein detection into a quantitative, sequencable readout. In essence, the antibodies are incubated with single-cell suspensions under conditions comparable to a conventional flow cytometry staining protocol, following which the antibody-derived DNA is amplified and sequenced alongside cDNA derived from cellular mRNA. A major advantage of CITE-seq is its capacity for simultaneous, high-throughput detection of protein markers and single-cell transcriptomes.
Limitless target detection
Because CITE-seq uses DNA-barcoded antibodies (available as TotalSeq™ reagents) rather than antibody-fluorophore conjugates to detect proteins of interest, the number of targets that can be analyzed in a single experiment is no longer constrained by the detection reagents. This allows for massive screening of cell-surface proteins, enabling researchers to generate increasingly data-rich information from sample material that is often available in only short supply.
Extending multiomics to bulk RNA sequencing studies
DNA-barcoded antibodies can also add value to bulk RNA sequencing by providing a complete proteogenomic profile of the sample. Bulk RNA sequencing studies are commonly used to analyze RNA expression from a population of cells in suspension but lack the resolution to detect more than a handful of proteins in one experiment. By staining suspension cells with TotalSeq™ antibodies prior to bulk RNA sequencing—BioLegend's adaptation of the CITE-seq workflow known as bulk epitope and nucleic acid sequencing (BEN-seq)—it is now possible to profile numerous cell surface proteins alongside mRNA expression.
To compare the performance of CITE-seq and BEN-seq, researchers have characterized Th1 polarized human peripheral blood mononuclear cells (PBMCs) using 280 TotalSeq™ antibodies. As well as identifying a unique molecular profile characterizing Th1 cells, their results showed a strong correlation between the two methods.
Utility for studying SARS-CoV-2
A further important utility of DNA-barcoded antibodies is the study of innate and adaptive immunity against SARS-CoV-2. Using a panel of over 190 TotalSeq™ antibodies to analyze plasma and PBMC samples from healthy and COVID-19 afflicted patients, researchers have been able to resolve a major immunological shift between mild and moderate infection.
Robust software is essential to multiomics workflows
Making sense of the vast quantities of data generated during a multiomics study requires a highly intuitive software package. As well as allowing protein and RNA expression data to be viewed separately, multiomics analysis software should support clustering to enable markers of interest to more easily be identified. It is also important that analyses can be conducted in a cloud-based environment, particularly in a world that has become increasingly accustomed to remote working.
Antibody cocktails have been developed to remove the need for optimization of individual antibodies and simplify the CITE-Seq workflow. BioLegend’s TotalSeq™-C Human Universal Cocktail has been designed to react with 130 unique cell surface antigens, including principal lineage antigens, and includes 7 isotype control antibodies, to aid in the multiomic characterization of immune cells. It was recognized by The Scientist as one of the top 10 innovations of 2020 and is complemented by the TotalSeq™-A Universal Cocktail comprising 154 primary antibodies and 9 isotype controls.
This article originally appeared on Biocompare.
 Login / Register
Login / Register 






Follow Us