When incorporated into the protein of interest, epitope tags make protein detection and identification very robust and easy. A large number of different epitope tags are now available for use. BioLegend epitope tag antibodies, peptide tags, and affinity gels are extremely specific, highly characterized and generate clear, quantitative results. These epitope tag reagents are suitable for a wide variety of applications including Western Blot, Immunoprecipitation, Protein Purification, Flow Cytometry, and Immunofluorescence Microscopy.
Epitope Tags
Detection and purification of proteins and peptides depend on specific antibodies. There are many commercially available antibodies however they do not cover all proteins, especially the novel proteins, proteins of unknown sequences and proteins or peptides of low antigenicity. Raising an antibody to a protein of interest can be quite time-consuming and expensive. A convenient solution for detection and purification of various proteins is offered by Epitope tags (also known as affinity or fusion tags). Epitope tagging is a common experimental technique by which a well characterized and specific sequence of amino acids called the “epitope” (recognized by a particular antibody) is combined with a protein of interest. Antibodies directed against these specific epitope tags can bind to the fusion proteins bearing the epitope, thus allowing the detection of these tagged proteins. These epitope tags allow detection and purification without disturbing the structure of the protein to which they are fused. Choice of tag depends upon many factors, and one of the most important considerations is the downstream analysis of the protein such as western blot analysis, immunoprecipitation, and affinity purification.
Advantages of Epitope Tagging
- Saves time as epitope tagging is quicker than a custom antibody preparation, which requires a few months.
- Single epitope tagging is also cheaper than production cost of an antigen specific antibody.
- Same epitope tag can be used for various proteins of different sizes.
- The epitope tag is composed of 3 to 14 amino acids and does not disrupt normal protein functions.
- The technique can be used to detect proteins that are very difficult to isolate and purify.
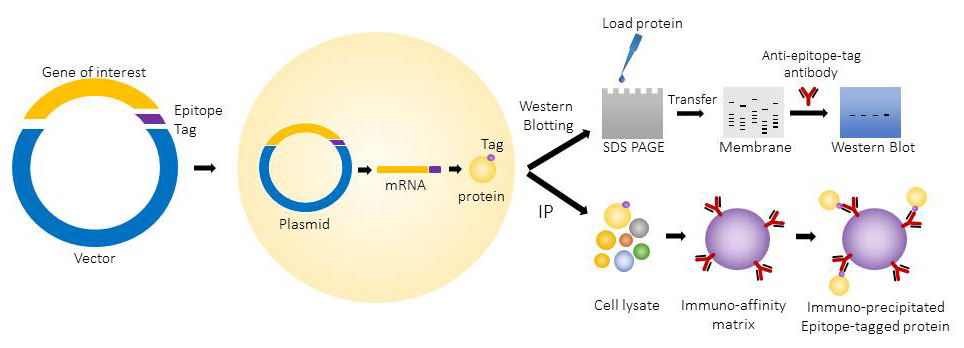
The epitope tagged proteins are commonly used for western blotting and immunoprecipitation. They can also be used for applications like protein purification, immunofluorescence microscopy and flow cytometry.
Affinity Gels and Matrices
Epitope tag antibodies can be conjugated to beads (agarose, sepharose, metal, etc.) in order facilitate isolation or immunoprecipitation of tagged proteins. BioLegend offers several affinity gels and matrices, including c-Myc, 6-His, DYKDDDDK, GST, GFP, and HA. These matrices can be re-used for multiple rounds of purification and display excellent specificity at an economical price.

SDS-PAGE gel image of protein purified from the 1st or 10th rounds by either BioLegend anti-DYKDDDDK agarose or a competitor’s equivalent product. Final lane contains ladder.

SDS-PAGE gel image of protein purified from the 1st to 10th rounds by BioLegend anti-c-Myc agarose. First and last lanes contain ladder. There is no loss of binding capacity even after ten rounds of re-use.
Epitope Tag Antibodies
The epitope tag antibodies we provide are extremely specific and provide outstanding performance for protein purification, Western blotting, and immunoprecipitation applications.
Purified anti-HA.11 Epitope Tag Antibody
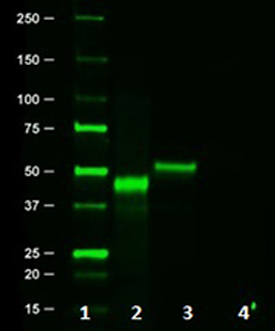
Western blot of Clone 16B12. Lane 1: Molecular weight marker; Lane 2: Posi-tag control Protein (Cat # 931301); Lane 3: HA.11 overexpressing cell lysate; Lane 4: Untransfected cell lysate.
Purified anti-DYKDDDDK Tag Antibody

Cell extracts from untransfected 293T cells (lane 1) or 293T cells transfected with a plasmid encoding DYKDDDDK-tagged protein (lane 2), using anti-DYKDDDDK, clone L5.
Purified anti-His Tag Antibody

Insect cell supernatant expressing His-tagged mouse CCL22 was resolved by electrophoresis, transferred to nitrocellulose, and probed with anti-His Tag antibody (clone J099B12). Proteins were visualized using a goat anti-mouse IgG secondary conjugated to HRP and chemiluminescence detection.
Recombinant Peptide Tags and Posi-Tag
BioLegend's Posi-Tag Epitope Tag Protein can be utilized as a positive control in Western blotting experiments where many epitope tags are commonly used. Posi-Tag represents proteins fused in the following order: GST, T7, HSV, c-Myc, VSV, Glu-Glu, V5, E-tag, DYKDDDDK, S-tag, HA, and 6-His, and has an approximate molecular weight of 45 kDa.
We also offer recombinant peptide tags that can be used during competition experiments to ensure specificity of your antibody. Alternatively, these synthetic peptides can be used to help elute the corresponding tagged protein from its affinity matrix.

Western blot showing several BioLegend epitope tag antibodies and their detection of 1 μg of Posi-Tag Epitope Tag Protein. All monoclonal antibodies were used at a 1:1000 dilution. Poly9248 was used at a dilution of 1:50000.
The AU1 epitope is represented by the amino acid sequence DTYRYI, derived from the major capsid protein of bovine papillomavirus-1 (BPV-1). It is a commonly used epitope tag engineered onto either the N- or C- terminus of a protein of interest so that the tagged protein can be analyzed and visualized by immunochemical methods.
Also derived from the major capsid protein of bovine papillomavirus-1 (BPV-1), the AU5 epitope is mapped to the amino acid sequence TDFYLK. AU5 is a very commonly used epitope tag that can be engineered onto the N- or C- terminus of a protein of interest so that the tagged protein can be analyzed and visualized by immunochemical methods. Due to the short sequence, this modification is not likely to affect the structure or function of the recombinant proteins.
Cre-recombinase is a 38kD DNA recombinase derived from the P1 bacteriophage and consists of 343 amino acids. It is highly specific for a 34 bp DNA sequence (loxP) found in P1 DNA. It requires no energy cofactors, such as ATP, to carry out its recombination function. The products of this recombination event depend on orientation and location of the loxP sites.
c-Myc tag (or Myc tag) originated from the c-myc proto-oncogene, cellular homolog of the avian v-myc gene in humans. It is only 10 amino acids long (EQKLISEEDL) and this small size tag does not perturb (or enhance) protein function. It can be fused to the C-terminus or the N-terminus of a protein, or even inserted within a protein. Multiple epitope tags can significantly enhance the sensitivity of detection of the fusion protein. This has been extensively used for western blotting, immunoprecipitation, and flow cytometry.
DYKDDDDK tag, commonly called Sigma's FLAG® tag, is the second fully functional epitope tag to be published in the scientific literature. Unlike other tags where a monoclonal antibody was first isolated against an existing protein, the FLAG® epitope was an idealized, artificial design to which monoclonal antibodies were raised. This has been extensively used for protein purification, western blotting, immunoprecipitation, and flow cytometry. To improve the detection of the FLAG® tag the 3x FLAG® system has been developed. This three- tandem FLAG® epitope is hydrophilic, 22-amino-acids long, and efficient for detection of low abundance proteins. If used at the N-terminus of the protein, the FLAG® tag can be removed by treatment with enterokinase, which is specific for the five N-terminal amino acids of the FLAG peptide.
Our reagent, clone NT73, can be utilized to detect E. coli RNA polymerase and is designed to recognize Softag 1. Softag 1 is an immunoaffinity system appropriate for purifying tagged proteins from eukaryotic expression sources. The epitope (SLAELLNAGLGGS) is derived from a subunit of prokaryotic RNAP.
Green fluorescent protein (GFP) is ~29 kDa fluorescent protein used as versatile marker for visualization of protein localization, monitoring physiological processes and for detecting transgenic or transient protein expression. GFP can be used for the generation of fusion proteins without significantly interfering with native protein assembly and function, which makes this tag a very powerful tool for in vivo analyses.
The Glu-Glu tag, CEEEEYMPME, was originally isolated from the polyoma virus medium T antigen and can be incorporated into the C- or N-terminus of a protein of interest. This tag is often used as a protein modification in order to simplify the labeling and detection of proteins. The short sequence of Glu-Glu is unlikely to affect the structure or function of modified proteins. BioLegend’s epitope tag antibody, clone Glu-Glu, is capable of recognizing both EYMPME and EFMPME sequences.
GST (glutathione S-transferase) was one of the first epitope tags to be used and is ~26 kDa in size. Single-step purification of polypeptides as fusions with GST was first described as early as 1988. It can be placed on the N or C-terminus and can enhance the solubility of expressed proteins. GST tag is relatively large compared with other common peptide based epitope tags, and it may interfere with some protein functions. In these cases, it may be easily removed by protease cleavage.
The HA tag is derived from the surface glycoprotein of human influenza hemagglutinin (HA) molecule corresponding to amino acids 98-106. This segment is chosen due to its high immunogenicity and immuno-inaccessibility in the native hemagglutinin conformation. The HA epitope, YPYDVPDYA, has been widely used as a tag in recombinant protein expression vectors. This tag does not appear to interfere with the bioactivity or the biodistribution of the recombinant protein. The tag facilitates the detection, isolation, and purification of recombinant proteins.
Also known as a polyhistidine or hexa histidine-tag, this tag consists of at least six histidine (His) residues in tandem. It is extensively used for purification of recombinant proteins due to the affinity of the tag towards several types of immobilized metal ions under specific buffer conditions, including nickel, cobalt, and copper. The His-tag sequence can be placed on the N- or C-terminus of a target protein by using commercially available cloning vectors. This tag is often followed by a cleavage site for a specific protease to remove the tag after protein purification.
The ~43-kDa maltose-binding protein (MBP) is encoded by the malE gene of E. coli K12. MBP is a monomeric protein. Usually, the protein of interest is expressed as a MBP-fusion protein. Once added to the recombinant protein, MBP can increase the solubility of over- expressed fusion proteins in bacteria, especially eukaryotic proteins. The mechanism by which MBP increases solubility is not well-understood. A spacer sequence coding for ten asparagine residues between the MBP and the protein of interest increases the chances that a particular fusion will bind tightly to the amylose resin. The MBP-tag can be easily detected using immunoblotting and immunoprecipitation. The MBP can be fused at the N- or C-terminus of the protein.
mCherry is a second generation fluorescent protein derived from DsRed, a red fluorescent protein related to GFP, isolated from disc corals of the genus Discosoma. The 'm' in the name refers to its monomer configuration. The prototype for such fluorescent proteins is Green Fluorescent Protein (GFP), which is a ~27 kDa protein isolated originally from the jellyfish Aequoria victoria. mCherry is resistant to photobleaching, is quite stable, and has an extremely rapid maturation rate.
Our reagent, clone IIB8, can be utilized to detect RNA polymerase TFIIB by recognizing Softag3. Softag 3 is an immunoaffinity system appropriate for purifying tagged proteins from prokaryotic expression sources. The epitope (TQDPSRVG) is derived from a subunit of eukaryotic transcription factor TFIIB.
S-tag or S-15 is an epitope tag derived from first 15 N-terminal amino acids, KETAAAKFERQHMDS, of the pancreatic ribonuclease A (RNase A). When the RNase A is digested with enzyme subtilisin, it yields two components, S-peptide and S protein, which remain together and have full enzymatic activity of the parent protein (now called RNase S). The separation of components abolishes enzymatic activity, but when they are mixed together again, fully active RNase S is re-formed. This unique property of reconstituting enzymatic activity by mixing the S tag peptide and S-protein fraction enables sensitive quantitative measurement of S-tagged fusion protein by a simple assay.
The Strep-tag is a selected eight amino acid peptide (WRHPQFGG) that has intrinsic binding affinity towards streptavidin and has been extensively used for purification for membrane embedded recombinant proteins. The tag can be placed at the C- or N-terminus. Due to its small size, the tag does not interfere with folding or bioactivity, and does not induce protein aggregation.
T7
The T7 epitope tag is composed of an 11-amino acid long peptide encoded from the leader sequence of the T7 bacteriophage gene 10. A major T7 capsid protein is encoded by gene 10, but its function is not clear. Due to the small size of the tag, it does not affect the tagged protein's biochemical properties. The tag is commonly engineered onto the N- or C- terminus of a protein of interest and is useful for the detection of proteins using immunoblotting, immunoprecipitation, and immunostaining techniques. The T7 tag has been used extensively as a general epitope tag and is commonly available in many commercial expression vectors including pET vectors.
Thioredoxin is an 11.7 kD E. coli cytoplasmic protein that act as an anti-oxidant in cells. When used as a fusion partner to produce recombinant protein, thioredoxin has been shown to increase protein solubility and yield, as well as reduce protein aggregation and precipitation.
The V5 tag is derived from the RNA polymerase α subunit of simian parainfluenza virus type 5 and corresponds to amino acids 95-108. The V5 epitope represents GKPIPNPLLGLDST and can be used to detect expression of recombinant proteins in bacteria, yeast, insects, and mammalian systems.
VSV-G
This tag is derived from the vesicular stomatitis virus (VSV), which is a negative-strand RNA virus. The VSV genome encodes for 5 proteins: N, P, M, G, and L. The G protein (glycoprotein) is located at the virion surface and is responsible for virus attachment and penetration. The VSV-G antibody recognizes the five C-terminal residues of the tag (YTDIEMNRLGK). Like many tags, it is commonly available as a cassette in commercially available vectors to aid cloning.
What are the advantages of Epitope tags?
- Tag can be easily and rapidly added to a known gene.
- Multiple tags can be added if required.
- Well-characterized antibodies are available.
- The antibody is specific to the tag, therefore cross-reaction with other proteins is avoided
- Proteins and protein complexes can be purified using standardized practices.
- Tagged proteins can be distinguished from otherwise identical untagged proteins
- Possible to study novel and poorly immunogenic proteins.
What are the limitations of Epitope tags?
A few can be as follows
- A cloned and characterized gene or cDNA must be available
- The epitope tag may interfere with protein structure or function
- Epitope-tagged genes can be expressed at abnormal levels due to the use of heterologous promoters
- The epitope-tagged gene must be introduced into the cell, tissue, or organism of interest.
How do I introduce the tag?
The two standard approaches to tagging a cloned gene are: (a) An epitope encoding oligonucleotide is inserted into the coding sequence, or (b) the coding sequence is inserted into an expression vector that already carries the epitope tag. BioLegend does not provide cloning vectors or oligos for cloning. The cloning protocols can be found online or in the literature. When tagging by oligonucleotide insertion, it is important to take into account codon usage preferences for the target cell or organism.
Will the epitope tag interfere with the function of my protein?
That has to be determined empirically. However, it is possible that the insertion of the epitope tag may interfere with protein function.
Where should I put the epitope tag?
In the literature, the tag has been placed at, or very near, the extreme N or C terminus of the target protein. There are a few reasons behind this choice. Some are historical-like the first proteins to be epitope tagged were tagged at the termini, and when new proteins were tagged it seemed wise to do what had worked in the past. Some of the reasons are practical-tagging is often performed using expression vectors that automatically put the tag at a terminus. And some reasons are theoretical-termini are frequently chosen in the belief that proteins will tolerate additions more readily at these locations than at other sites. While the latter belief may be true-termini are rarely included in active sites, for example-it is also true that many proteins are known for which terminal sequences are critical for function. The termini also appear favorable because they are likely to be on the outside of the folded polypeptide, where one wants the tag to go, and not in the hydrophobic core. But it must be remembered that, due to simple geometry, most of the amino acids in any protein are on the outside, and so, if a protein is tagged at a randomly chosen site, the tag will probably wind up on the outside anyway.
Why is there no signal with the epitope tag antibodies on a western blot?
A lack of signal following western blotting may indicate a few different problems.
- No or very poor transfer. This can be addressed by quickly checking the membrane with Ponceau-S staining.
- The expression level of the GFP-tagged protein may be too low. Load more ug quantity of the sample and include a positive control.
- A very diluted antibody may be the problem, try a few different concentrations of the antibody to probe the western blot.
- Also, a remote possibility is that the GFP tag is out of frame and is not expressed resulting in a lack of any signal.
Proteins can be detected with an anti-protein antibody, but not with the epitope tag antibody. What's happening here?
It is possible the target protein was not tagged, the tag is out of the reading frame, or the protein is degraded.
How to set up controls for co-immunoprecipitation experiment?
For negative control, use an unrelated antibody, or control with the same host species, class, and subclass as the immunoprecipitating antibody. For positive control, use an expression vector with only the epitope tag of interest.
During Western blot with epitope tag antibodies, there are multiple bands.
Several reasons can account for this:
- the early termination of the translation of epitope-tagged protein;
- non-specific detection with anti-epitope tag antibody due to no/poor blocking of the immunoblot.
- partial degradation of the protein
- the concentration of the primary or the secondary antibody is too high.
- Washing between the incubations is not efficient
What applications can I use epitope tagged antibodies for?
The commonly used applications are western blotting, immunoprecipitation and protein purification. These can also be used for immunofluorescence microscopy and flow cytometry.
 Login / Register
Login / Register 







Follow Us