A Deeper View of Cancer with Single-Cell Multiomics
Cancer cells exist in complex populations, and cells within a single tumor exhibit molecular and phenotypic variations. Single-cell omics applications have proven to be effective in identifying these variations and informing the development of precision medicines. These applications, like RNA or
DNA sequencing, provide a comprehensive dataset describing the transcriptome, genome, proteome, or other areas of biology. Advanced multiomics applications have transformed cancer research by providing a method to integrate these datasets and link changes in gene expression or DNA mutations to protein expression.
We created oligonucleotide-conjugated TotalSeq™ antibodies and reagents for highly-multiplexed single-cell protein detection that integrate seamlessly into a variety of multiomics applications. Explore the capabilities of TotalSeq™ reagents.
See how researchers are using our TotalSeq™ reagents to accelerate cancer research by determining mechanisms of regulation of immune checkpoint inhibitors and creating an atlas of human tumors.
Identification of Immune Checkpoint Regulatory Pathways

Immune checkpoint molecules, like PD-L1, negatively regulate T cell activation and stimulate tumor escape and survival. Understanding how these molecules are upregulated on cancer cells provides insights for the development of new immunotherapies.
In a recent Nature Genetics article, Papalexi, et al. studied the mechanisms and molecular networks that upregulate PD-L1 in cancer cells. Using an emerging multiomics application called ECCITE-seq (expanded CRISPR-compatible cellular indexing of transcriptomes and epitopes by sequencing), the authors combined pooled CRISPR screens with single-cell mRNA and surface protein detection to simultaneously investigate transcriptional and post-transcriptional regulation of immune checkpoints. This technique expands on previous technologies by capturing single guide RNAs that deliver the CAS9 protein for CRISPR-mediated genetic knockouts. First, using CITE-seq in stimulated and unstimulated cells, they identified well-characterized and putative regulators of PD-L1. Next, they used pooled CRISPR libraries targeting these regulators and performed ECCITE-seq, pairing changes in gene and protein expression with each genetic knockout.
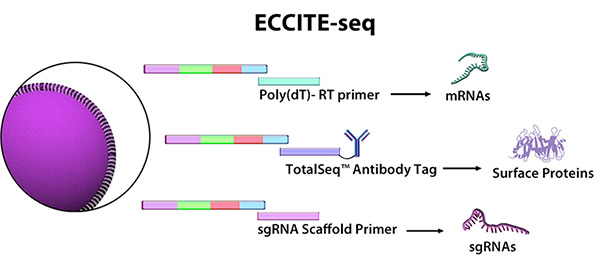
ECCITE-seq, developed by the New York Genome Center, combines single-cell mRNA, surface protein, and sgRNA detection.
The authors identified 8 regulators of PD-L1, including CUL3, MYC, and BRD4 as negative regulators. Interestingly, knockout of CUL3 affected both protein and mRNA expression of PD-L1. CUL3 is part of a ubiquitin ligase complex that degrades proteins, indicating there may be an indirect mechanism by which CUL3 regulates PD-L1 mRNA levels. Further transcriptional analysis demonstrated that the CUL3-containing ubiquitin ligase degrades NRF2 protein. NRF2 modulates PD-L1 gene expression by directly binding to the promoter. Therefore, CUL3 is capable of directly and indirectly affecting PD-L1 expression.
These studies highlight the advantages of multiomic characterization by linking transcriptional and post-transcriptional readouts to reveal new insights into PD-L1 regulation.
Creating a Single-Cell Atlas of Human Breast Cancers
Like many tumors, breast cancers are highly heterogenous. Understanding the cell types comprising different tumor subtypes and how these cells interact with the microenvironment enhances our knowledge of disease pathology and may lead to the development of personalized therapies.

See how Wu et. al. used CITE-seq for in-depth immunophenotyping in this Nature Genetics publication. They performed surface protein and transcriptional profiling of breast cancer cells from 26 tumors across multiple cancer subtypes. Integrating these datasets, the authors created a high-resolution cellular taxonomy describing 9 major cell types, including epithelial, immune, and stromal cells, representing 29 to 49 different cellular states. Phenotyping of the immune cells revealed novel PD-L1/PD-L2+ macrophages, extending previous work that correlated this novel cell type with disease progression3, 4. Within the stromal microenvironment, the authors identified 3 major cell populations of 3-5 identifiable states. Complementary spatial analysis examining the interactions of these cells with the host environment supported the idea that mesenchymal cell populations were separated, indicating that their differentiation or migration may depend on signals from the tumor microenvironment. Using cell surface markers identified in this atlas, these cells can be isolated and further characterized.
Together, this study demonstrates how complementary methods, including multiomics applications, can provide a cellular atlas for continued study and understanding of complex populations, like breast cancer. Using this data to grouping tumors with similar cellular makeups may provide new insights into disease pathology, subtype characterization, and our knowledge of how to treat these cancers.
Our Solutions for Multiomics Applications
TotalSeq™ oligo-conjugated reagents enable protein detection by sequencing and integrate seamlessly into existing single-cell transcriptomics, bulk sequencing, or single-cell genomics workflows. To accelerate multiomics applications, our scientists created individual antibodies, pre-optimized cocktails, hashtag reagents for multiplexing, and free analysis software. Learn more about our products.
Single-cell multiomics is transforming cancer research and the development of precision therapies. Discover other ways our reagents are being used to accelerate discoveries in high-impact publications.
References
- Papalexi, E., Mimitou, E.P., Butler, A.W. et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat Genet 53, 322–331 (2021). https://doi.org/10.1038/s41588-021-00778-2
- Wu, S.Z., Al-Eryani, G., Roden, D.L. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 53, 1334–1347 (2021). https://doi.org/10.1038/s41588-021-00911-1
- Wagner, J. et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330–1345.e18 (2019).
- Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698.e14 (2019).
 Login / Register
Login / Register 






Follow Us