ChIP Troubleshooting and Optimization
 |
| Chromatin immunoprecipitation (ChIP) is an important application to help determine where transcription factors bind or identify epigenetic changes at a gene of interest. As with most applications, understanding and using the right protocol is critical for success. But, when that protocol doesn’t work, it's hard to know where things have gone wrong. Here, we'll discuss 3 major areas to optimize or troubleshoot your ChIP experiments. 1. Chromatin Quality The quality of the chromatin going into the IP is one of the most important aspects of a ChIP assay. Chromatin can degrade quickly, so ensure you always keep your samples on ice, and don't forget to add protease inhibitors during cell lysis. Once you've crosslinked and lysed your cells, there are two widely-used techniques for shearing chromatin: sonication and enzymatic digestion. |
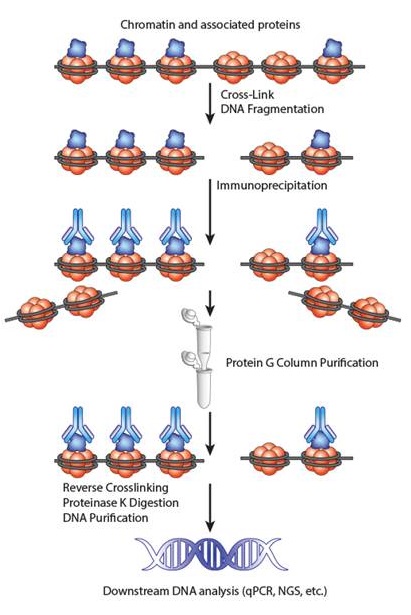 Overview of a ChIP experiment. |
|
| So, which method is ideal for your experiments? This may depend on what protein you are pulling down or what cells you are using. If you're looking at an abundant, tightly bound protein like a histone, then either method should work well. If you are looking at a transcription factor, it may be better to use a gentler method like enzymatic digestion or try sonication using a lower concentration of detergent in the buffer. Cell types that are hard to lyse may require harsher sonication-based shearing protocols. The exact conditions for shearing will depend on the specific cells and proteins you are analyzing. We'd recommend performing optimization experiments testing a variety of conditions before starting your IPs. Treat your cells exactly as you would for your experiment and run the resultant DNA on an agarose gel and look for shearing between 150-900 bp. With some cell types, even one extra cycle of sonication can cause over-shearing of your chromatin. |
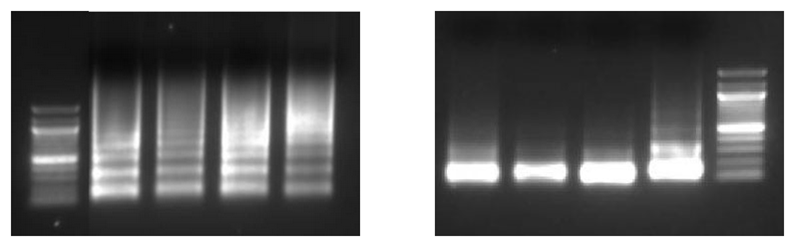 Chromatin from cell lines digested using MNase. On the left, chromatin was sheared optimally. On the right, chromatin was over-digested resulting in the majority of fragments below 400 bp. This can impair antibody binding resulting in little or no enrichment. |
| 2. Immunoprecipitation |
| Once you’ve prepared the chromatin, the next step is to immunoprecipitate your protein of interest. Setting up the IP is a fairly simple process but there are a few things to keep in mind at this step. |
|
| 3. Controls |
| Using the right controls allows you to properly analyze your downstream data at the end of the experiment. If something goes wrong, looking at how your controls performed will help you understand where to focus your troubleshooting efforts. Here, we'll outline the most important controls and how to use them. |
|
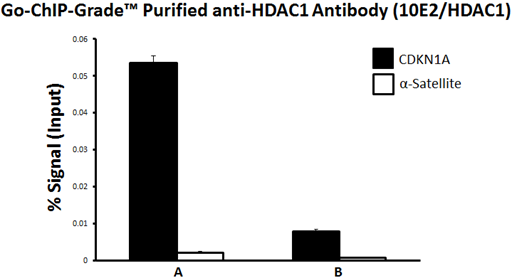 Looking at ChIP data, you can see how each of the controls help analyze the data. Here we used an anti-HDAC1 antibody (A) or an isotype control antibody (B). For each, we measured enrichment at a positive control locus, CDKN1A, and a negative control gene locus, an α-Satellite region. Lastly, by using an input control, we presented the enrichment relative to signal from the input control, so that we can directly compare samples IP'ed with two different antibodies. |
|
|
| Optimizing and troubleshooting your ChIP experiment can seem overwhelming, but breaking the experiment down can help understand where and why the experiment has failed. For more tips and tricks to help optimize your experiment, check out our ChIP webpage or watch our ChIP protocol video. |
| Contributed by Kelsey Swartz, Ph.D. |
 Login / Register
Login / Register 






Follow Us