
Infection or tissue injury causes innate immune cells such as Dendritic cells, Macrophages, and Neutrophils, to recognize pathogen-associated molecular patterns (PAMPs) or host derived damage-associated molcular patterns (DAMPs) via pattern recognition receptors (PRRs) on the cell surface and in the cytoplasm. The major PRRs on cells include Toll-Like receptors (TLRs), NOD-like receptors (NLRs), and retinoic acid-inducible gene 1 (RIG-I) like receptors (RLRs). BioLegend offers variety of reagents to study these pathways for innate immune cell signaling.
Toll-Like Receptor Signaling
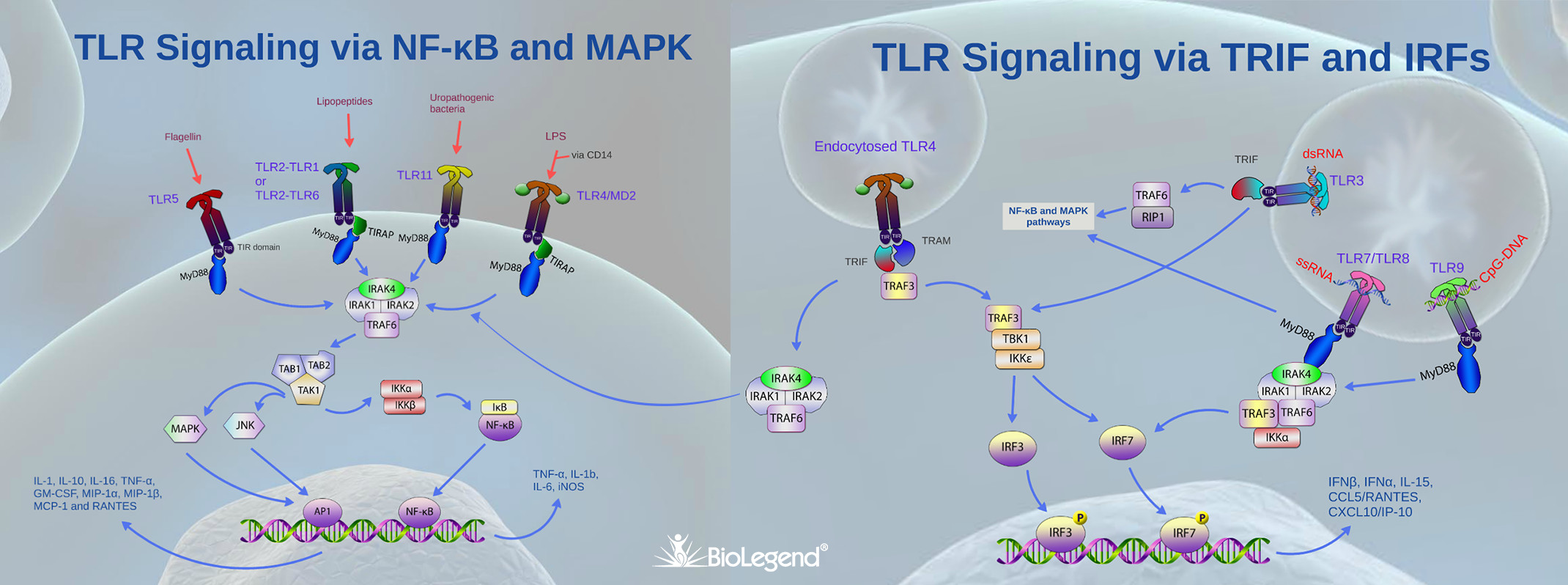
TLRs are the best studied family of PRRs, which recognize PAMPs of diverse pathogens including bacteria (Gram-positive and Gram-negative), DNA and RNA viruses, parasites, protozoa, and fungi. TLRs are broadly subdivided based on their location at the cell surface (these are activated by lipid and protein ligands) and endosomal compartments (these are activated by non-self nucleic acids).
TLRs typically dimerize upon ligand binding and their cytoplasmic domain called Toll/IL-1 (TIR, due to similarities to the IL-1 receptor) then serves as a docking site for TIR containing adaptor proteins. Out of several intracellular TIR containing proteins such as TRIF, TIRAP and TRAM, MyD88 acts a central player in innate immune signaling by being the canonical adaptor for inflammatory signaling pathways downstream of TLRs (except TLR3, which signals via TRIF), as well as IL-1 receptor families. MyD88 binds directly to the majority of TLR TIRs but in some cases the binding occurs in combination with adaptor TIRAP. MyD88 then recruits IRAK1 and IRAK4, which are later dissociated after being phosphorylated. The MyD88–IRAK complex also interacts with ubiquitin ligase TRAF6 to make polyubiquitin chains that activate the IKK complex for NF-κB and MAPK dependent gene transcription and production of pro-inflammatory cytokines such as IL-1β, IL-6, IL-8, and IL-12, and TNF-α. In addition, via the activation of interferon regulatory factor (IRF), MyD88 can also stimulate type 1 IFN genes leading to the production of antiviral interferons such as IFN-α , IFN-β, and IFN-λ
To view a step-by-step guide of the TLR signaling pathways, start the presentation below. (Best viewed in full screen mode. Use the [ ] icon at the bottom right.)
NLRs & AIM2
NOD-like-receptors play a key role in the recognition of cytosolic microbial and danger molecules. NLRs are sub-classified into five categories: NLRA, NLRB, NLRC, NLRP, and NLRX. The domain architecture of NLRs typically includes an N-terminal protein-protein interaction domain , caspase recruitment domain (CARD), pyrin domain (PYD), a central NACHT domain that helps with self-oligomerization, and a C-terminal leucine rich repeat (LRR) to detect PAMPs. NOD1 and NOD2 are well studied NLRs that recognize PAMPs. NOD1 is expressed in both hematopietic and non-hematopoietic cells and is activated by γ-D-glutamyl-meso-diaminopimelic acid (meso-DAP) from all Gram-negative bacteria and some Gram-positive bacteria. NOD2 is largely expressed in hematopoietic cells, certain epithelial cells and small Paneth cells. NOD2 signaling is triggered by cytosolic muramyl dipeptide found in the peptidoglycan of all bacteria. Stimulation of NOD1 or NOD2 by their ligands results in the formation of multi-protein signaling complex called the nodosome. The nodosome initiates the activation of the canonical NF-κB pathway that culminates with the production of pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6, IL-8, and RANTES.
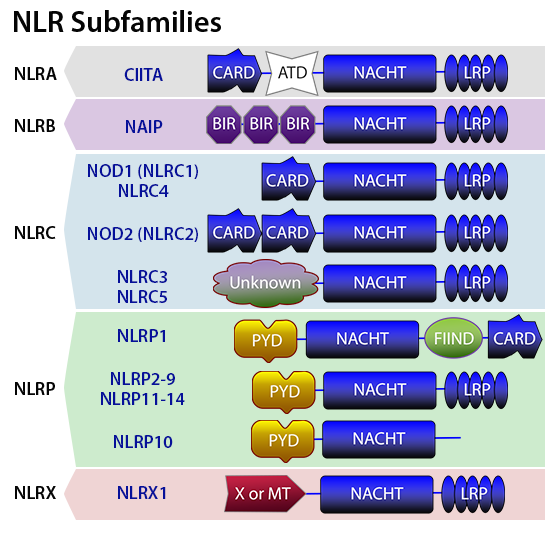
While NOD1 and NOD2 engage in the formation of nodosome, other NLRs contribute to the formation of macromolecular structure called the inflammasome. Both hematopoietic and non-hematopoietic lineages of cells exhibit the formation of inflammasome. The NLRP3 inflammasome is the most commonly studied among the NLRs. Typically, the inflammasome consists of an NLRP scaffold, the inflammatory protease caspase-1, and apoptosis associated spec like protein (ASC) adaptor, which upon activation triggers the maturation of pro-inflammatory cytokines such as IL-1β and IL-18 by proteolytic activation of caspase-1. Several DAMPs and PAMPs, such as viral and bacterial RNA and DNA, particulates (silica, asbestos and alum), ATP, flagellin, and Anthrax toxin, can cause inflammasome activation.
Foreign (viral and bacterial), as well as aberrant host double-stranded DNA recognition, is performed by a non-NLR protein called AIM2(absent in melanoma 2) using its HIN200 domain attached to a pyrin domain (PYD), which helps with ASC recruitment for inflammasone formation. Activation of the AIM2 inflammasone also leads to the maturation of proinflammatory cytokines IL-1β and IL-18.
To view a step-by-step guide of the NLR and AIM2 signaling pathways, start the presentation below. (Best viewed in full screen mode. Use the [ ] icon in the bottom right.)
Retinoic acid-inducible gene I (RIG-I) mediated signaling
Cellular antiviral signaling is initiated by the recognition of viral nucleic acids by cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including RIG-I, which is the most well studied. RIG-I contains a central DECH (DExD/H) box helicase domain for RNA binding and ATP hydrolysis, a C-terminal domain (CTD) for RIG-I autoregulation, and a CARD domain at the N-terminus. Viral RNA binding causes RIG-I derepression and dimerization. Lysine-63 (K63)-linked polyubiqutination by TRIM25, an E3 ubiquitin ligase, causes RIG-I activation and oligomerization, and its subsequent binding to adaptor mitochondrial IPS-1/MAVS. This leads to MAVS activation, which involves its aggregation into large polymers. These polymers in turn activate TRAF6 and TRAF3 leading to the activation of kinases IKK and TBK1, which in turn activate NF-κB and IRF3. These transcription factors initiate the production of type I IFNs and inflammatory cytokines such as IL-1β and IL-18.
To view a step-by-step guide of the RIG-I signaling pathway, start the presentation below. (Best viewed in full screen mode. Use the [ ] icon at the bottom right.)
 Login / Register
Login / Register 







Follow Us