- Clone
- 2F3 (See other available formats)
- Regulatory Status
- RUO
- Other Names
- H2A.x, H2a/x, Histone 2A, Histone 2A.X, Gamma-H2AX
- Isotype
- Mouse IgG1, κ
- Ave. Rating
- Submit a Review
- Product Citations
- 97 publications

| Cat # | Size | Price | Quantity Check Availability | Save | ||
|---|---|---|---|---|---|---|
| 613401 | 25 µg | 95€ | ||||
| 613402 | 100 µg | 235€ | ||||
H2A.X is a 14 kD basal histone and a member of the H2 histone family. This nuclear protein is synthesized in the G1 and S phase of the cell cycle and is known to be important for DNA repair and maintaining genomic stability and for recombination between immunoglobulin switch regions. H2A.X becomes phosphorylated on serine 139 after double-stranded DNA breaks. Phosphorylated H2A.X promotes DNA repair and maintains genomic stability. The 2F3 monoclonal antibody reacts with phosphorylated human H2A.X (Ser139) and has been shown to be useful for Western blotting, immunofluorescence and flow cytometry.
Product DetailsProduct Details
- Verified Reactivity
- Human, Mouse
- Antibody Type
- Monoclonal
- Host Species
- Mouse
- Immunogen
- Modified peptide
- Formulation
- This H2A.X antibody is provided in phosphate-buffered solution, pH 7.2, containing 0.09% sodium azide. Final antibody concentration is 0.5 mg/ml.
- Preparation
- The antibody was purified by affinity chromatography.
- Concentration
- 0.5 mg/ml
- Storage & Handling
- Upon receipt, store between 2°C and 8°C.
- Application
-
WB - Quality tested
ICC - Verified
ICFC - Reported in the literature, not verified in house - Recommended Usage
-
Each lot of this antibody is quality control tested by Western blotting. Western blotting, suggested working dilution(s): Use 5 µg antibody per 5 ml antibody dilution buffer for each mini-gel. For immunocytochemistry, a concentration range of 1.0 - 4.0 μg/ml is recommended. It is recommended that the reagent be titrated for optimal performance for each application.
- Application Notes
-
Additional reported applications (for the relevant formats of this clone) include: immunohistochemistry on paraffin embedded sections2, immunofluorescence microscopy3-9, Western blotting 10-12, and flow cytometry1,13. Clone 2F3 cross-reacts with mouse4.
Intracellular staining protocol for Anti-H2A.X-Phosphorylated (Ser139) Antibody for Flow Cytometry
1. Prepare 70% absolute ethanol. Chill solution by storing at -20°C.
2. Prepare cells of interest.
3. Wash 1X with PBS, centrifuge at 350g for 5 min.
4. Discard the supernatant and vortex to loosen cell pellet.
5. Add pre-cooled 70% ethanol drop by drop, while vortexing.
6. Incubate at -20°C for 60 minutes.
7. Wash 3X with BioLegend Cell Staining Buffer and resuspend the cells at 0.5-1 X 107 cells/ml in the cell staining buffer.
8. Perform immunofluorescent staining for flow cytometry. -
Application References
(PubMed link indicates BioLegend citation) -
- Jha JC, et al. 2013. J. Virol. 87:5255. (FC) PubMed
- Akbay A, et al. 2008. Am J Pathol. 173:536. (IHC) PubMed
- Mochizuki K, et al.2008.J cell Sci.121:2148. (IF) PubMed
- Xiao R, et al. 2007. Mol Cell Biol.27:5393. (IF) PubMed
- Rossi DJ, et al. 2007. Nature. 447:725. (IF) PubMed
- Loidl J, et al. 2009. Mol Cell Biol. 20:2048. (IF) PubMed
- Beels L, et al. 2009. Circulation. 120:1903. (IF) PubMed
- Suzuki K, et al. 2010. Nucleic Acids Res. 38:e129. (IF) PubMed
- Lukaszewicz A. 2010. Chromasoma Apr 27. [Epub ahead of print] (IF) PubMed
- Yamada C, et al. 2010 J. Biol. Chem. 285:16693. (WB) PubMed
- Bu Y, et al. 2010, Biochem Biophys Res Commun. 397:157. (WB) PubMed
- Massignan T, et al. 2010. J. Biol Chem. 285:7752. (WB) PubMed
- Banath JP, et al. 2010. BMC Cancer 10:4 (FC)
- Zhang M., et al. 2011. Cancer Res. 23:7155. PubMed
- Kuefner MA, et al. 2012. Radiology 264:59. PubMed
- Yoshihara Y, et al. 2012. Biochem Biophys Res Cmmun. 421:57. PubMed
- Titus S, et al. 2013. Sci Transl Med. 13:21. PubMed
- Crown KN, et al. 2013. G3. 6:1927. PubMed
- Schenkwein D, et al. 2013. Nucleic Acids Res. 41:e61. PubMed
- Zhadanova NS, et al. 2014. Mol Cell Biol. 34:2786. PubMed
- Horrell SA, et al. 2014. Eukaryot Cell. 13:1300. PubMed
- Maya-Mendoza A, et al. 2015. Mol Oncol. 9:601. PubMed
- Product Citations
-
- RRID
-
AB_315794 (BioLegend Cat. No. 613401)
AB_315795 (BioLegend Cat. No. 613402)
Antigen Details
- Structure
- Basal histone, H2 histone family; 14 kD
- Distribution
-
Nuclear
- Function
- Phosphorylated H2AX promotes DNA repair and maintains genomic stability. Important for recombination between immunoglobulin switch regions
- Modification
- Phosphorylation on Ser139 after double-stranded DNA breaks
- Biology Area
- Cell Biology, Chromatin Remodeling/Epigenetics, DNA Repair/Replication, Neuroscience
- Molecular Family
- Phospho-Proteins
- Antigen References
-
1. Mannironi C, et al.1989. Nucleic Acids Res. 17:9113.
2. Celeste A, et al. 2002. Science 296:922.
3. Bassing CH, et al. 2002. Proc. Natl. Acad. Sci. USA 99:8173.
4. Reina-San-Martin B, et al. 2003. J. Exp. Med. 197:1767. - Regulation
- Synthesized in G1 and S-phase of cell cycle
- Gene ID
- 3014 View all products for this Gene ID
- UniProt
- View information about H2A.X Phospho Ser139 on UniProt.org
Related Pages & Pathways
Pages
Related FAQs
Other Formats
View All H2A.X Phospho (Ser139) Reagents Request Custom Conjugation| Description | Clone | Applications |
|---|---|---|
| Purified anti-H2A.X Phospho (Ser139) | 2F3 | WB,ICC,ICFC |
| FITC anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
| Alexa Fluor® 488 anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
| Alexa Fluor® 647 anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
| Alexa Fluor® 594 anti-H2A.X Phospho (Ser139) | 2F3 | ICC |
| PE anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
| PerCP/Cyanine5.5 anti-H2A.X-Phosphorylated (Ser139) | 2F3 | ICFC |
| Direct-Blot™ HRP anti-H2A.X-Phosphorylated (Ser139) | 2F3 | WB |
| APC anti-H2A.X-Phosphorylated (Ser139) | 2F3 | ICFC |
| PE/Cyanine7 anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
| APC/Fire™ 750 anti-H2A.X Phospho (Ser139) | 2F3 | ICFC |
Customers Also Purchased
Compare Data Across All Formats
This data display is provided for general comparisons between formats.
Your actual data may vary due to variations in samples, target cells, instruments and their settings, staining conditions, and other factors.
If you need assistance with selecting the best format contact our expert technical support team.
-
Purified anti-H2A.X Phospho (Ser139)
Total lysates (15 µg protein) from untreated HeLa (Lane 1) a... 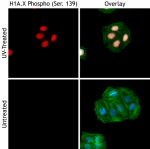
HeLa cells were stained with purified anti-H2A.X Phospho (Se... -
FITC anti-H2A.X Phospho (Ser139)
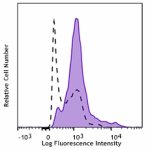
HeLa cells untreated (open histogram) or treated with Nocoda... -
Alexa Fluor® 488 anti-H2A.X Phospho (Ser139)
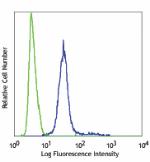
Nocodazole-treated Hela cells intracellularly stained with 2... -
Alexa Fluor® 647 anti-H2A.X Phospho (Ser139)
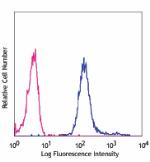
Nocodazole-treated Hela cells intracellularly stained with 2... -
Alexa Fluor® 594 anti-H2A.X Phospho (Ser139)
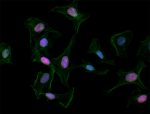
HeLa cells were fixed with 1% paraformaldehyde (PFA) for ten... -
PE anti-H2A.X Phospho (Ser139)
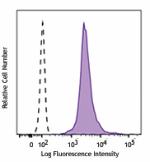
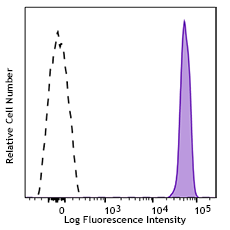
Nocodozole treated HeLa cells (24 hours) were fixed and perm... -
PerCP/Cyanine5.5 anti-H2A.X-Phosphorylated (Ser139)
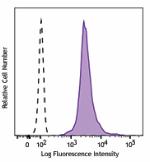
Nocodozole treated HeLa cells (24 hours) were fixed and perm... -
Direct-Blot™ HRP anti-H2A.X-Phosphorylated (Ser139)
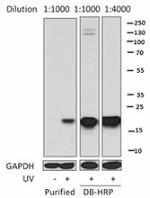
15 µg of total protein extracts from UV untreated and treate... -
APC anti-H2A.X-Phosphorylated (Ser139)
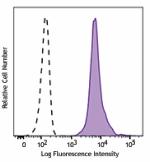
Nocodazole treated HeLa cells (24 hours) were fixed and perm... -
PE/Cyanine7 anti-H2A.X Phospho (Ser139)
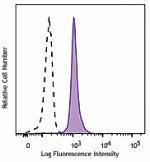
Nocodazole treated HeLa cells (24 hours) were fixed and perm... -
APC/Fire™ 750 anti-H2A.X Phospho (Ser139)
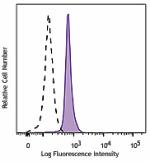
Nocodazole treated HeLa cells (24 hours) were fixed and perm...

 Login / Register
Login / Register 
















Follow Us