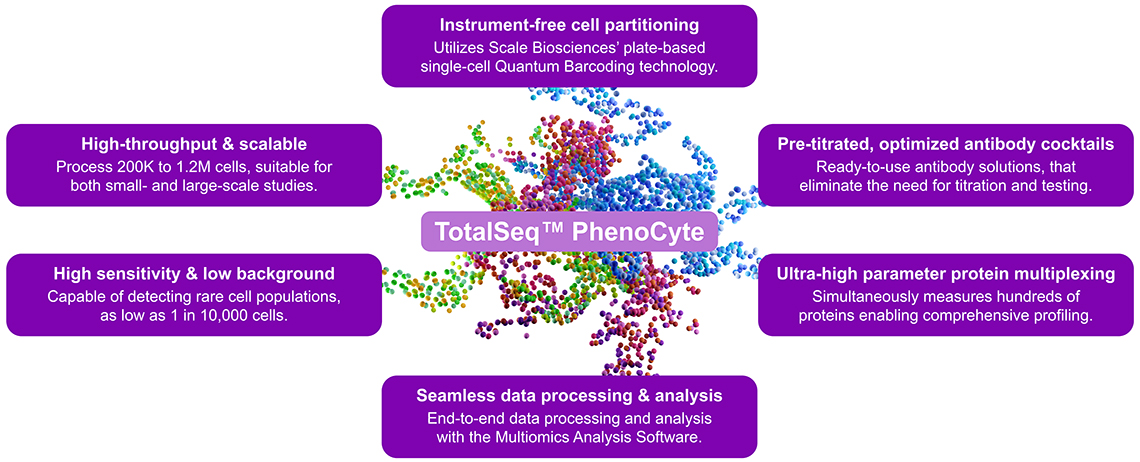
Overall Workflow
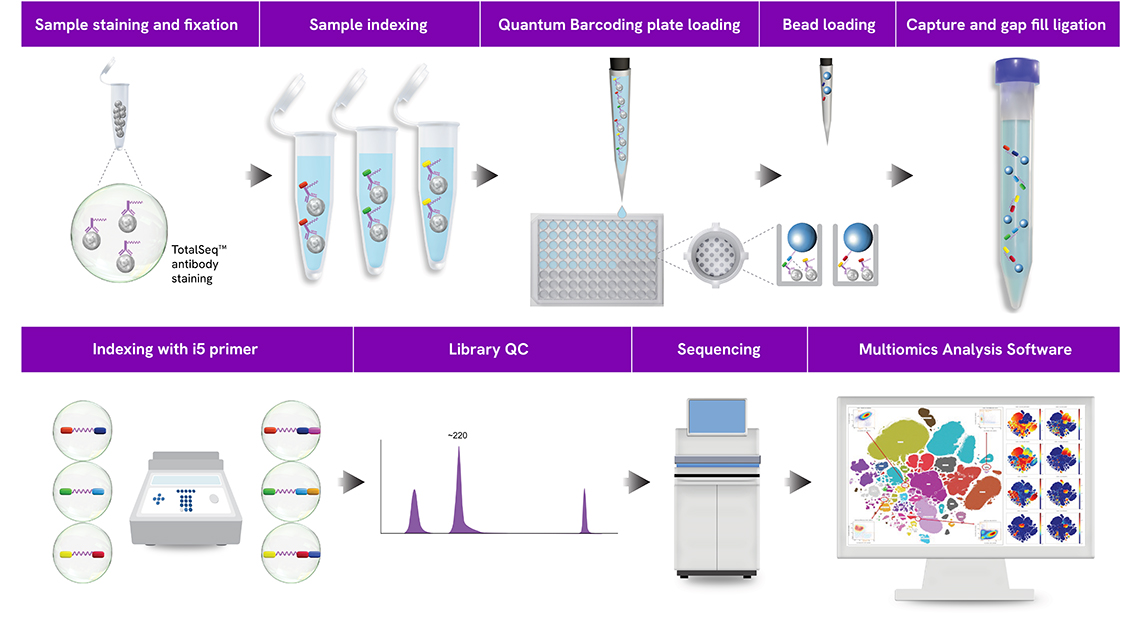
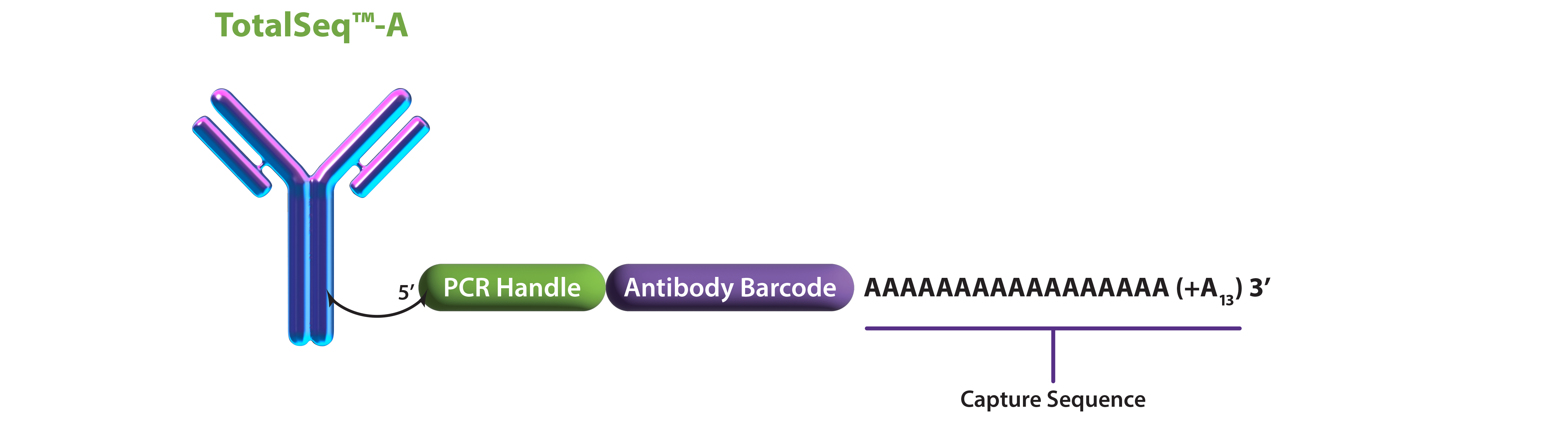
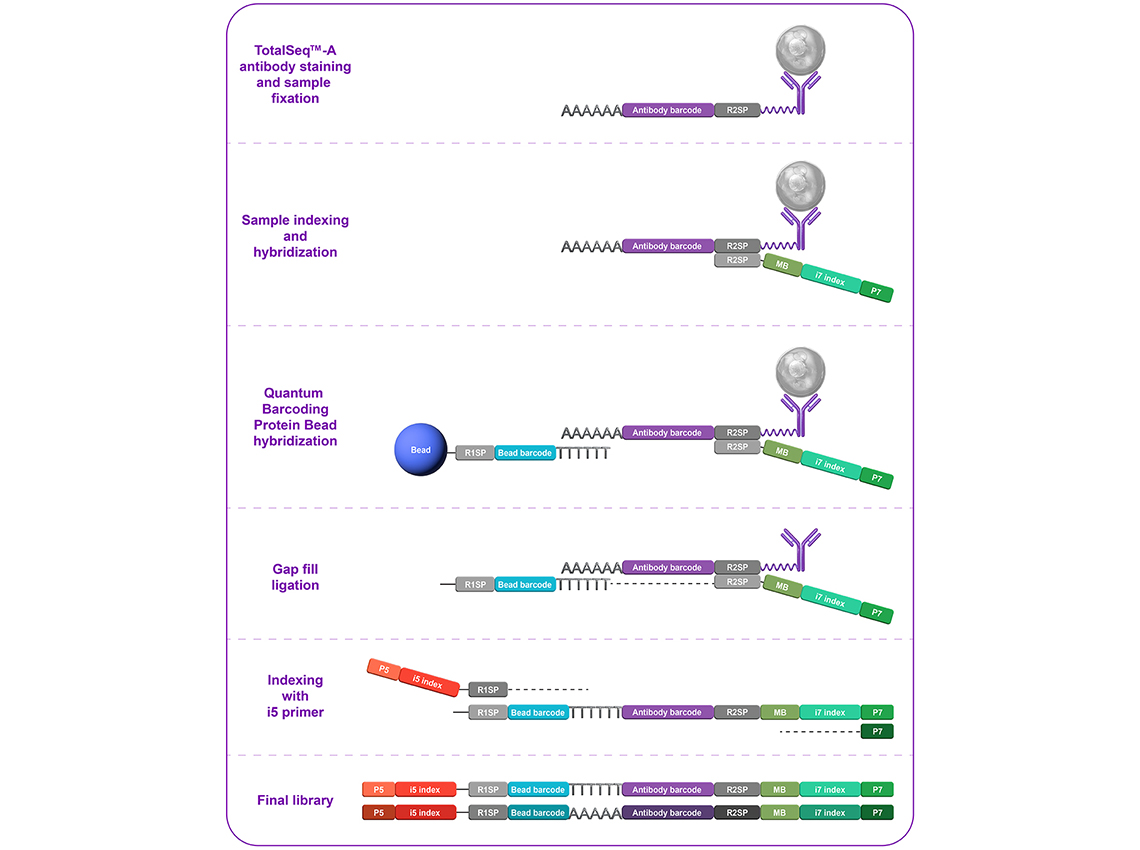
Sample staining and fixation: Cells stained with TotalSeq antibodies and subsequently fixed with paraformaldehyde.
Sample indexing (i7 primer): Cells are partitioned and indexed with various Sample Indexing Oligos (i7 primers) which hybridize to the antibody oligos.
Quantum Barcoding plate loading: Sample indexed (i7) cells are loaded onto the Quantum Barcoding plate that partition cells into microwells.
Bead loading: Quantum Barcoding protein beads containing the bead barcode (second index) are loaded onto the plate and hybridize directly to the poly(A) tail of the antibody oligos.
Capture and gap fill ligation: Post-hybridization, the beads now containing the hybridized sample index (i7) and bead barcodes are collected and undergo a gap-fill ligation to bridge the indexing sequences.
Indexing with i5 primer: The ligated product undergoes a final round of indexing via i5 indexing PCR and the library is amplified.
Library QC: Determine library size and quantification.
Sequencing: A final library consists of combinations of unique identifiers: the Sample Index, the antibody barcode, the bead barcode, and the i5 index. The identifiers help facilitate the sequencing of the libraries and with the identification of individual cells in the downstream analysis.
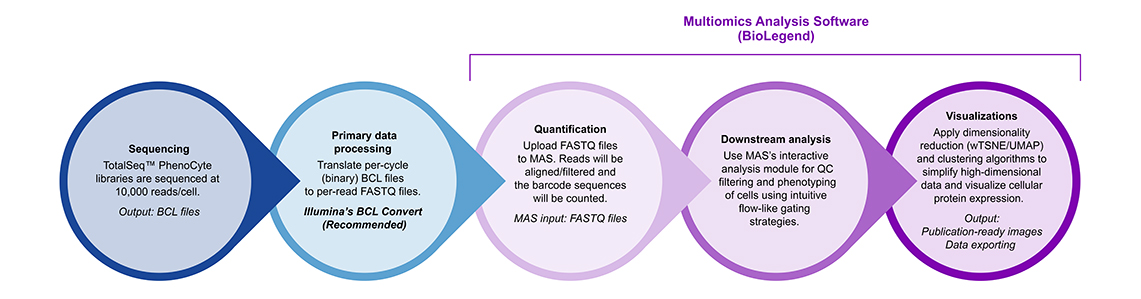
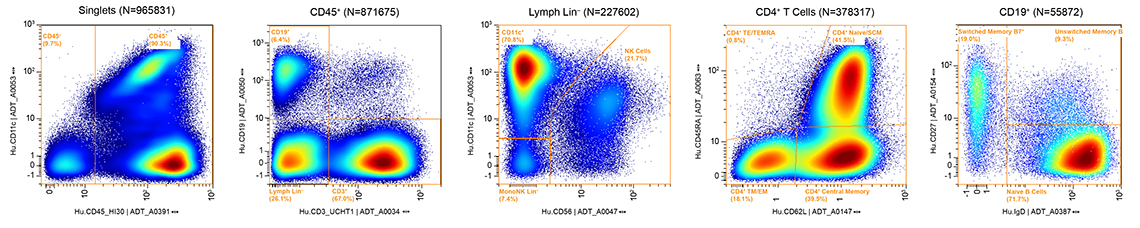
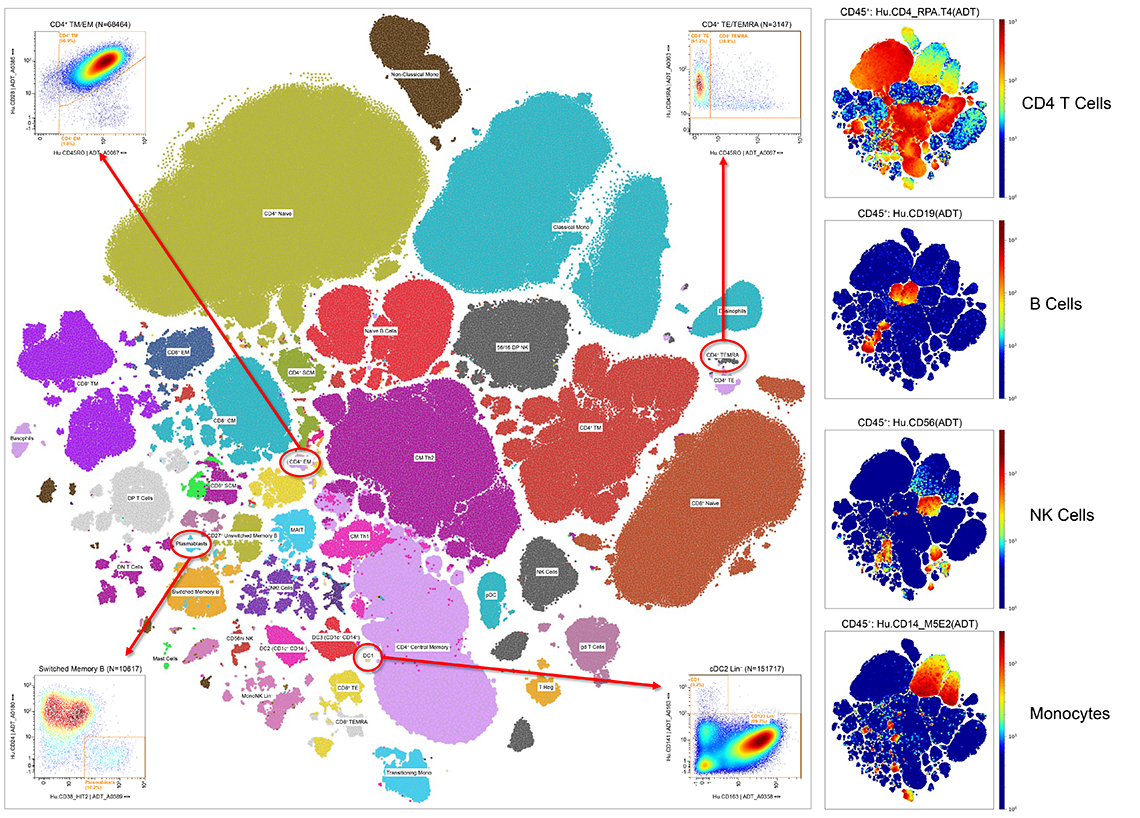
Multiomics Analysis Software: Data can be processed and analyzed with our free cloud-based Multiomics Analysis Software.
Note: A final library consists of combinations of three unique identifiers: the i7 index, the bead barcode, and the i5 index, which facilitate the identification of individual cells in the downstream analysis.

 Login / Register
Login / Register 








Follow Us