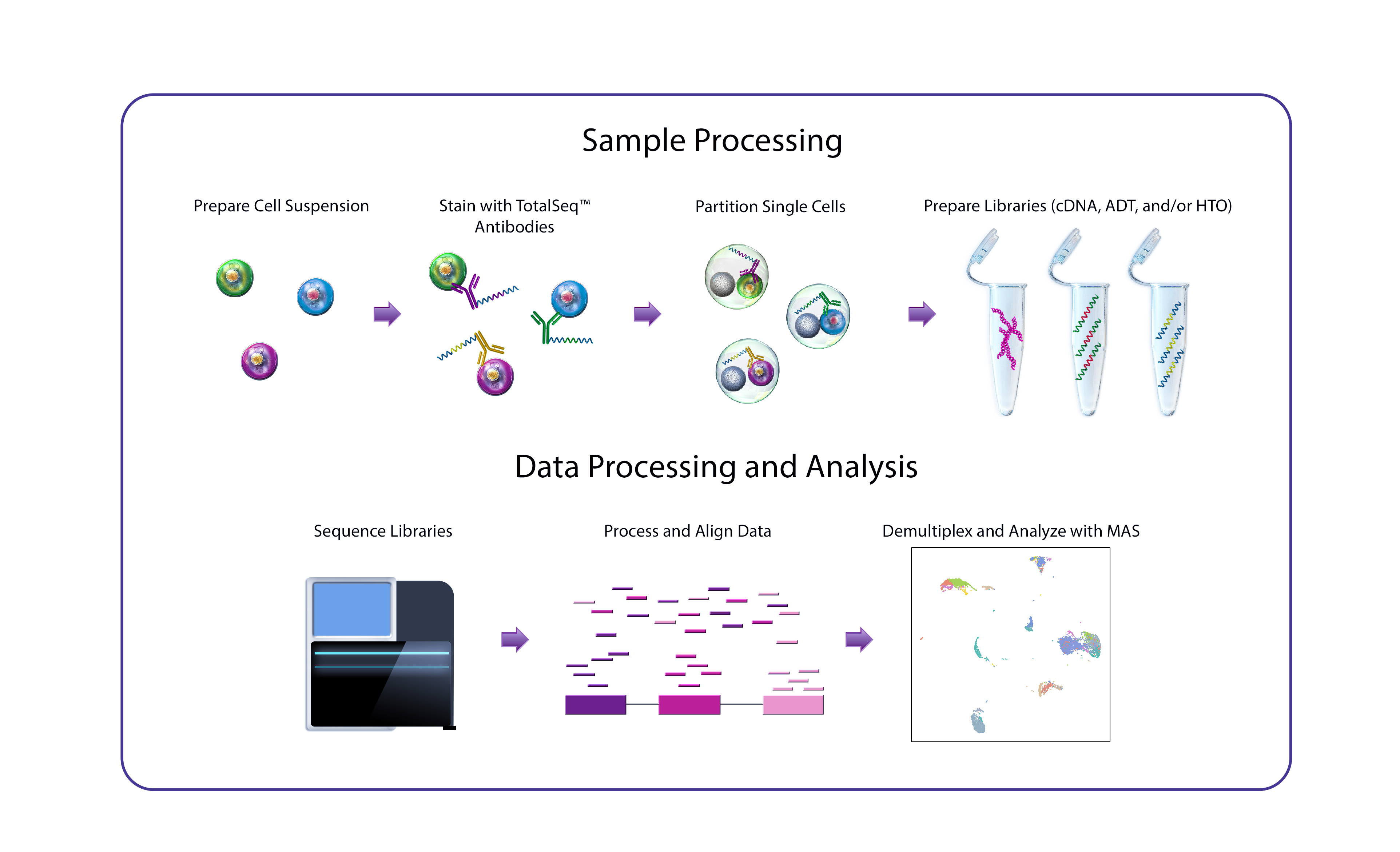
Single-Cell Protein and RNA
オリゴヌクレオチド標識抗体TotalSeq™を使用して、1細胞レベルでのタンパク質発現を解析し、ユニークな表現型を発見しましょう。これらの試薬は既存の1細胞シーケンス法に円滑に統合することができ、タンパク質とRNAを同時に解析することが可能となります。ここでは、プロテオゲノミクス解析で何ができるのか、そして、精密医療や腫瘍学、免疫学、神経科学や幹細胞研究などにおける新規アプリケーションで、TotalSeq™試薬がどのようにマルチプレックス法を可能とするのかを見てみましょう。
TotalSeq™フォーマットは、BioLegend独自の特許出願中の技術です。この技術に関して、当社との提携にご興味をお持ちの方はこちらからお問い合わせください。
TotalSeq™ Reagents for Single-Cell Protein and RNA Detection
TotalSeq™ oligo-conjugated antibodies enable measurement of proteins at a single-cell level and integrate seamlessly into existing single-cell RNA sequencing workflows, including Drop-Seq and those available from 10x Genomics.
Simultaneous Multiomic Data Generation: Increase the power of single-cell experiments by combining proteomic and transcriptomic data.
Reduced Dropouts: In contrast to mRNA, TotalSeq™-derived antibody tags are not highly prone to dropouts, which are essentially false negative readouts.
Ultra-High Parameter Surface and Intracellular Protein Detection: Detect hundreds of proteins in a single cell. A lower dropout rate contributes to enhanced sample clustering and the ability to better identify specific cell types.
Diverse applications: With a wide-range of mouse and human targets available, you can use TotalSeq™ antibodies in a variety of research areas, including:
- Personalized or Precision Medicine
- Cancer Research
- Stem Cell Research
- Basic and Applied Immunology
- Biomarker Discovery
- Characterization of New or Rare Cell Types
- Neuroimmunology
- Vaccine Research
Explore Additional Resources
Visit our 10x Genomics partnership page to learn how our TotalSeq reagents integrate with their single-cell solutions.
Access our optimized protocols for staining of surface and intracellular proteins specifically developed for use with our TotalSeq reagents for CITE-Seq applications. Explore our full list of TotalSeq antibodies for surface proteins and intracellular proteins.
For more information regarding the development of the CITE-Seq methodology, check out the CITE-Seq website.
Discover how our TotalSeq™ reagents and recombinant proteins can be used to build antigen multimers to characterize B cell response to infection with LIBRA-seq.
Highlighted Publications
Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19
Su Y. et al. Cell. 2020; October. DOI: 10.1016/j.cell.2020.10.037
A major study aimed at performing multiomic analyses to characterize immune responses from COVID-19 patients. Using a panel of over 190 TotalSeq™ antibodies, this study identified novel immune cell populations that emerge in patients experiencing moderate COVID-19 severity that are expanded in severe cases. Additionally, the study identified changes within plasma metabolite profiles of patients experiencing severe COVID-19 disease that indicate that depletion of certain metabolites is correlated with the development of severe disease.
A Conserved Dendritic Cell Regulatory Program Limits Antitumour Immunity
Maier B, et al. Nature. 2020; doi: 10.1038/s41586-020-2134-y.
Immune checkpoint blockade is a recent cancer treatment method that induces a durable antitumor response. This therapy aims to mitigate the tolerogenic immune response induced by dendritic cells upon presentation of tumor antigen to T cells. However, it is only effective in a limited subset of cancer patients. This study aims to understand the mechanism by which enhancement of systemic antitumor T cell immunity occurs after neoadjuvant PD-L1 blockade. Through the use of single-cell proteogenomics, the authors identify a dendritic cell regulatory program that limits antitumor activity and is induced by expression of IL-4. Blockade of IL-4 results in increased antitumor responses, suggesting a potential treatment for the improvement of immune checkpoint therapy.
Discover Antibody-Oligo Formats
Each TotalSeq™ antibody is conjugated to a unique oligonucleotide containing a capture sequence, a clone-specific barcode sequence, and a PCR handle compatible with Illumina® sequencing reagents and instruments. Barcodes are placed between the PCR handle and 3’ flanking sequence. The oligonucleotide sequence that is conjugated to our TotalSeq™ antibodies is also referred to as an Antibody-Derived Tag, or ADT. The oligo sequence that is conjugated to our hashtag reagents is referred to as a Hashtag Oligonucleotide, or HTO.
For single-cell protein and RNA analysis, we offer TotalSeq™-A, -B, and –C formats and continue to work with compatible partners to support new versions of genomics reagents and solutions.
Understand the Differences Between TotalSeq™ Formats
TotalSeq™-A: Contain a poly (A) capture sequence that mimics a natural mRNA. Designed to work with platforms that utilize poly (dT) oligonucleotides as the mRNA capture method. Compatible with 10x Genomics 3' Universal Gene Expression assay, captured via the poly (dT) sequence of the Gel Bead primers. Use of TotalSeq-A reagents with this assay requires reagent and protocol modifications; 10x Genomics provides only limited support for TotalSeq-A. For additional information regarding the use of TotalSeq-A reagents, please refer to our protocols or contact BioLegend Technical Support.
TotalSeq™-B: The capture sequence is compatible with the Capture Sequence 1, or Partial Capture Sequence 1 of the barcoded Gel Bead primers used with the 10x Genomics 3’ Universal Gene Expression or Flex Gene Expression assays, respectively. Use of TotalSeq-B antibodies with these assays is fully supported by 10x Genomics.
TotalSeq™-C: The capture sequence is compatible with the Template Switch Oligo Capture Sequence, or the Partial Capture Sequence 1 of the barcoded Gel Bead primers used with the 10x Genomics 5' Universal Gene Expression or Flex Gene Expression assays, respectively. The use of TotalSeq-C antibodies with these assays is fully supported by 10x Genomics.
For more information regarding compatibility of our TotalSeq products with partner solutions, such as 10x Genomics, please visit our Technology Partnerships and Service Providers page.
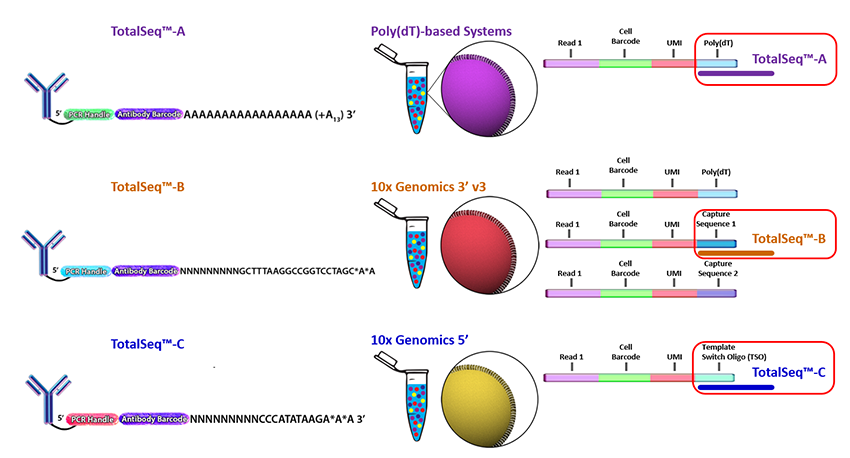
10x Genomics' Solutions, compatible with TotalSeq™-B and -C antibodies, contain all necessary reagents to process and amplify oligos derived from both TotalSeq™ reagents and mRNA. When using TotalSeq™-A reagents with the 10x Genomics' Single Cell 3' Universal Gene Expression assay, additional reagents for preparation of libraries must be purchased. Please refer to our protocols for a complete list.
| TotalSeq™-A | TotalSeq™-B | TotalSeq™-C | |
|---|---|---|---|
| 10x Genomics compatibility | Chromium Single Cell Universal 3' Gene Expression assay and any system employing poly (A) tail capture method | Chromium Single Cell Universal 3' Gene Expression and Flex Gene Expression assays | Chromium Single Cell Universal 5' Gene Expression and Flex Gene Expression assays |
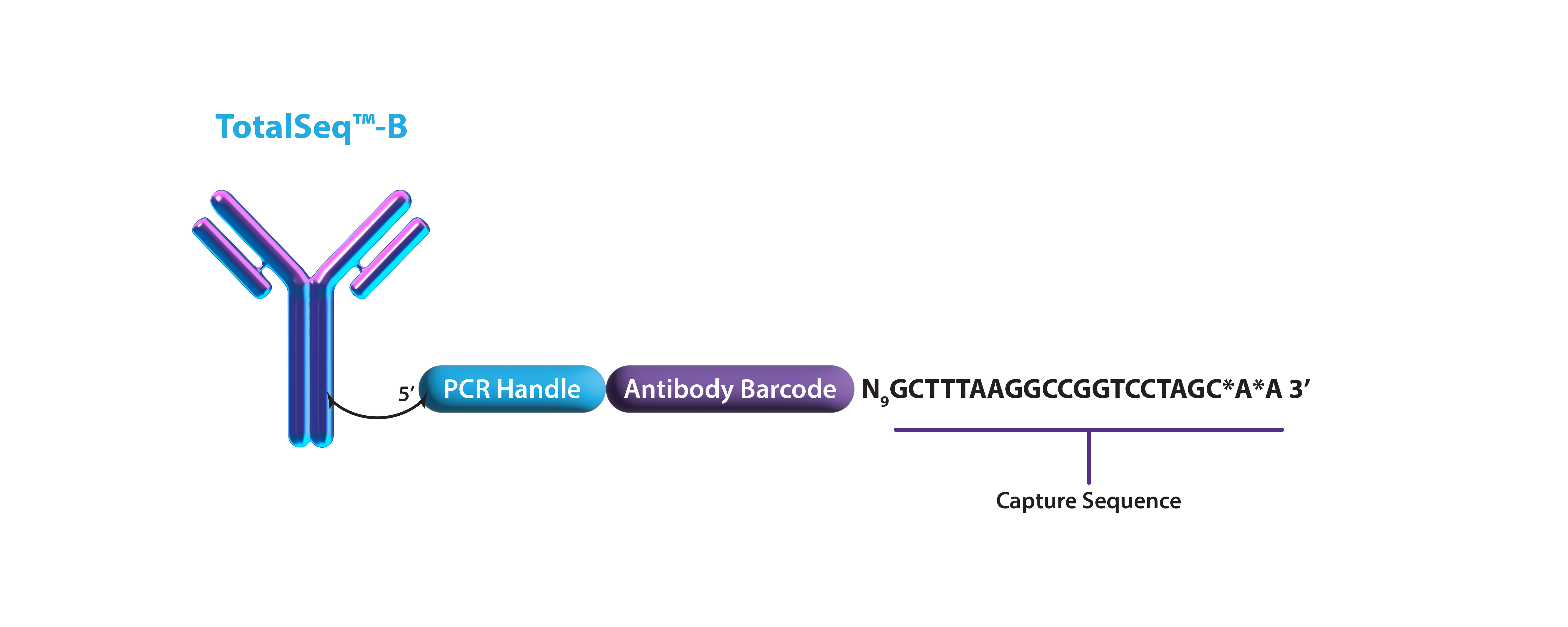
| PCR handle | CCTTGGCACCCGAGAATTCCA | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNN2 | CGGAGATGTGTATAAGAGACAGNNNNNNNNNN |
| Capture sequence | Poly-A [(A)30*A*A3] | NNNNNNNNNGCTTTAAGGCCGGTCCTAGC*A*A4 | NNNNNNNNNCCCATATAAGA*A*A4 |
| Next-generation sequencing compatibility | Compatible with Illumina instruments | Compatible with Illumina instruments | Compatible with Illumina instruments |
Notes:
- N represents a randomly selected A, C, G, or T.
- The symbol * indicates a phosphorothioated bond, to prevent nuclease degradation.
- These sequences are unique to the TotalSeq™-B and -C conjugates, and were developed independently of the reagents used by researchers at the New York Genome Center (NYGC, CITE-seq.com). TotalSeq™-B, -C, and the antibodies used by NYGC are all compatible with 10x Genomics' Solutions, but utilize different oligo sequences. As such, protocols and additional reagents, including required primers, differ between the antibody formats. Please refer to our protocols when using TotalSeq™ reagents.
The DepleteX® Single Cell Boost Kit can be paired with CITE-Seq single-cell workflows that utilize TotalSeq™ oligo-conjugated antibodies from BioLegend for protein detection.
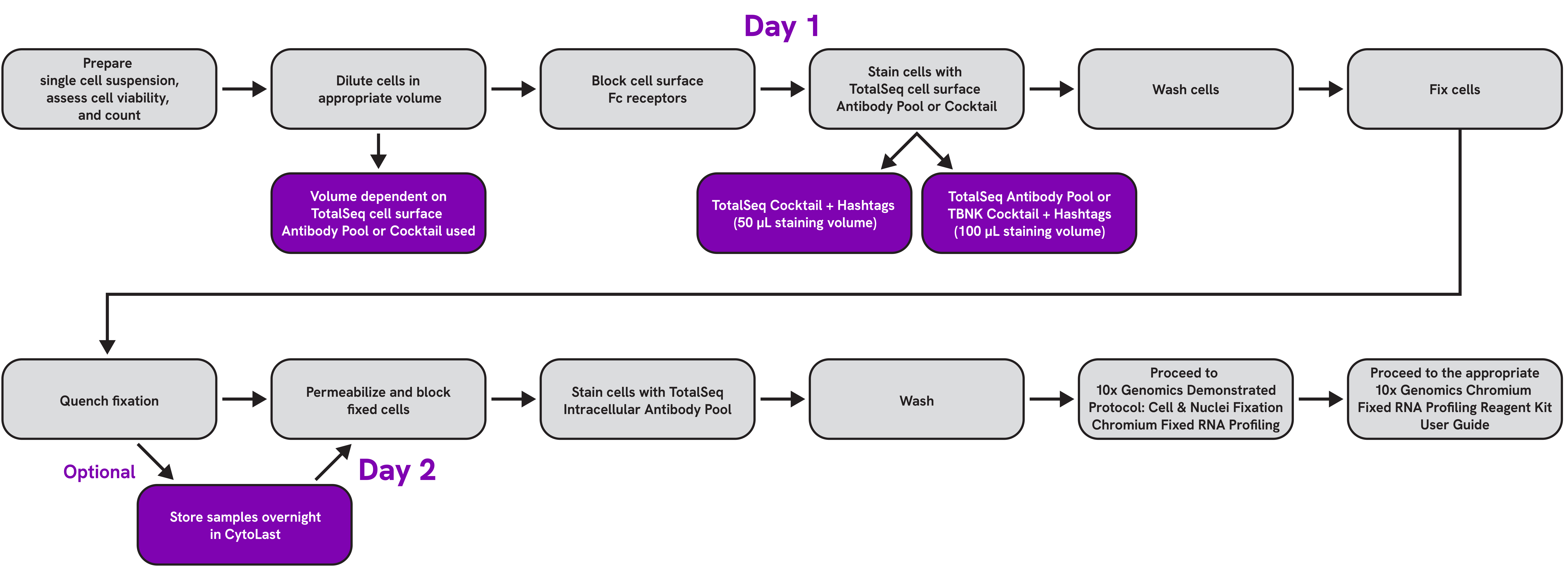
TotalSeq™-B and -C Reagents for Intracellular Cytoplasmic and Cell Surface Staining
Intracellular proteins contribute to a host of cellar functions including cell growth, gene regulation, chemotaxis, and immunomodulation. Measuring both intracellular and surface proteins at the single-cell level can give researchers a comprehensive understanding of the cell and the molecular processes that underlie cellular function.
The ability to detect cell surface proteins with gene expression in single-cell analysis has been well established using CITE-Seq. However, detecting intracellular proteins while preserving RNA quality for gene expression profiling has proven challenging, due largely to the conditions utilized to access the intracellular compartment of cells which can have deleterious effects on RNA quality.
With advancements in single-cell RNA technology and workflow optimization, it is now possible to observe all 3 modalities using TotalSeq antibodies and our TotalSeq™-B or -C Cell Surface and Intracellular Cytoplasmic Protein Staining with 10x Chromium Single Cell Gene Expression Flex.
Why add intracellular targets to your single-cell experiment?
- Improve cell characterization by identifying functional molecules.
- Add depth to cell state identification by understanding signaling pathways.
- Discover unique and new phenotypes within seemingly homogeneous cell populations.
Our full protocol compatible with surface protein expression is now available.
TotalSeq™ Format Compatibility

Assay Compatibility
|
TotalSeq Format Compatibility |
TotalSeq-B Surface and Intracellular Antibodies |
TotalSeq-C Surface and Intracellular Antibodies |
|
Single Cell Assay Compatibility |
||
|
BioLegend TotalSeq Intracellular Staining Protocol |
||
|
Instrument Compatibility |
10x Genomics Chromium X Series |
|
|
Description |
Probe based whole transcriptome gene expression, compatible with fresh or fixed samples |
|
|
Additional Cellular Features |
Surface and Intracellular Proteins |
|
|
Multiplexing Compatibility |
TotalSeq-B Hashtags |
TotalSeq-C Hashtags |
*Use of the TotalSeq-B or -C Cell Surface and Intracellular Cytoplasmic Protein Staining with 10x Chromium Single Cell Gene Expression Flex Protocol and our cell hashing reagents with 10x Single Cell Gene Expression Flex is not officially supported by 10x Genomics. For additional information regarding use of TotalSeq-B and -C intracellular antibodies or TotalSeq Hashtags please contact BioLegend Technical Support.
Workflow
Required User Guides and Support
|
TotalSeq Format |
TotalSeq-B |
TotalSeq-C |
|
BioLegend TotalSeq™ Intracellular Staining Protocol |
||
|
10x User Guides |
||
|
10x General Support |
||
Important:
*TotalSeq-B products are only compatible with 10x Genomics Chromium Fixed RNA Profiling Reagent Kits for Singleplexed Samples With Feature Barcode technology for Protein.
**TotalSeq-C products can be used with GEM-X Flex Gene Expression Reagent Kits for Singleplex or Multiplex samples with Feature Barcode technology for Protein Expression, and Next GEM Fixed RNA Profiling Reagent Kits for Singleplexed or Multiplexed Samples with Feature Barcode technology for Protein using Barcode Oligo.
For additional support in choosing the correct Flex Gene Expression user guide to pair with our TotalSeq products please contact BioLegend Technical Support.
TotalSeq Reagents
TotalSeq-B: The capture sequence is compatible with the Capture Sequence 1, or Partial Capture Sequence 1 of the barcoded Gel Bead primers used with the 10x Genomics 3' Universal Gene Expression or Flex Gene Expression assays, respectively. Use of TotalSeq-B antibodies with these assays is fully supported by 10x Genomics.
TotalSeq-C: The capture sequence is compatible with the Template Switch Oligo Capture Sequence, or the Partial Capture Sequence 1 of the barcoded Gel Bead primers used with the 10x Genomics 5' Universal Gene Expression or Flex Gene Expression assays, respectively. The use of TotalSeq-C antibodies with these assays is fully supported by 10x Genomics.
|
TotalSeq Intracellular Antibodies |
||
|
Cocktails |
||
|
Hashtags |
||
|
All Products |
Additional TotalSeq Resources
TotalSeq Resources and Learning - View all our TotalSeq resources including training videos, protocols, and app notes.
Barcode Look-Up - Access our barcode look-up table to view and export barcode information for all of our TotalSeq reagents.
Intracellular and Surface Staining Representative Data
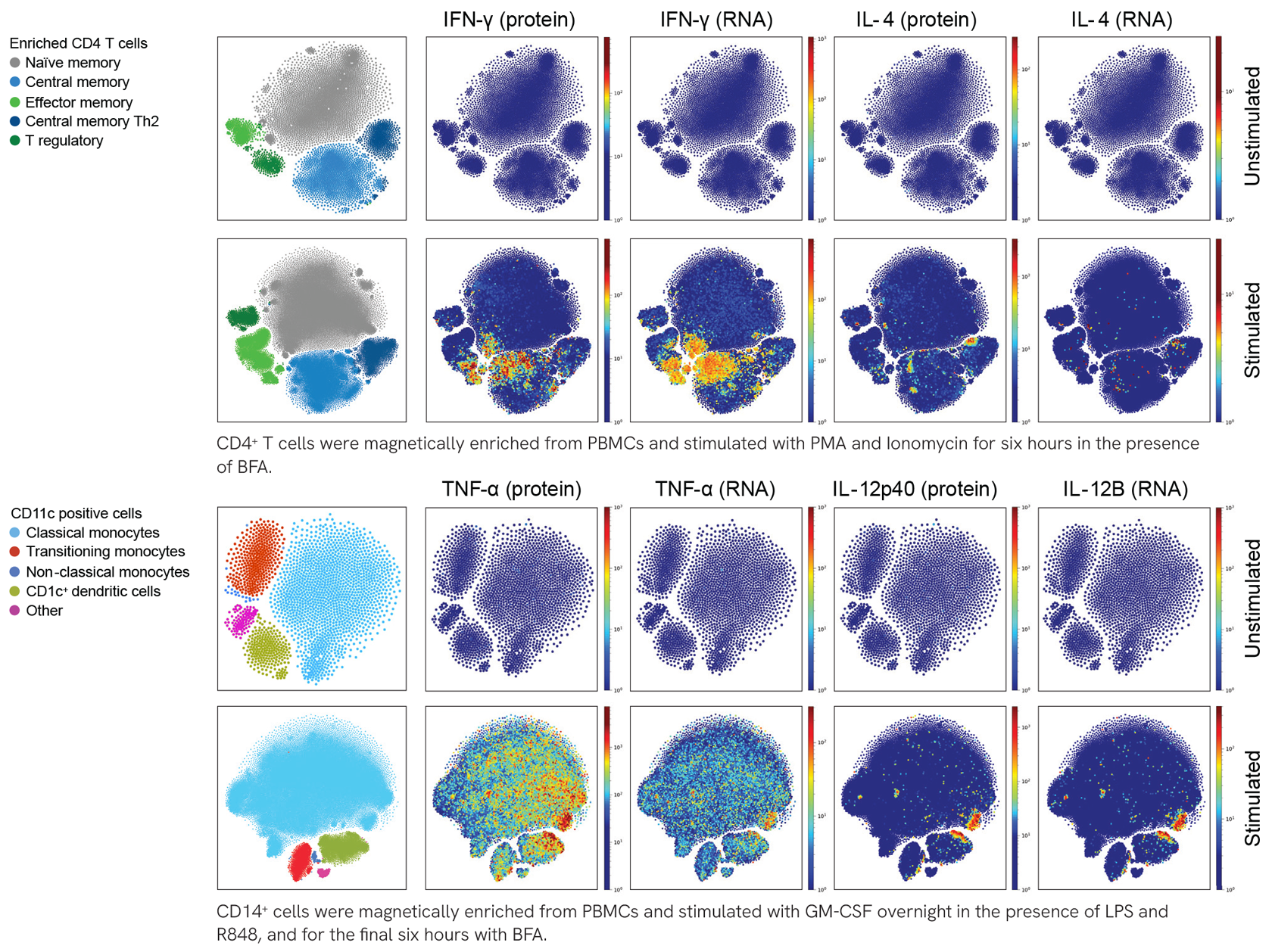
Multiplex Samples with Hashtag Antibodies
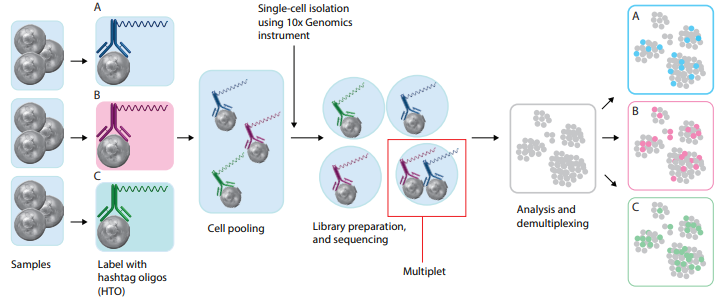
Cell Hashing
To pool multiple samples prior to loading them onto a platform capable of single-cell isolation, try one of our hashtag reagents. Each pre-mixed, ready-to-use hashtag reagent contains a pool of antibodies designed to recognize ubiquitously expressed cell surface markers and is conjugated to a unique barcode. For human samples, our hashtag reagents recognize CD298 and β2-Microglobulin, and for mouse samples, hashtag antibodies recognize CD45 and H-2 MHC Class I.
Benefits of using cell hashing:
- Robust multiplet identification.
- Minimize variability and reduce batch effects between samples.
- Pool smaller samples to reach minimal cell numbers.
Read more about cell hashing and how they can be used to multiplex samples in the initial paper by Stoeckius M, et al.
Tech Insights: Cell Hashing with TotalSeq™ Reagents
Multiplexing samples in single-cell experiments improves sample throughout and provides cost savings. Learn from Nathan Lucas, PhD, as he describes the benefits and technical considerations for our TotalSeq™ hashtag reagents.
Nuclei Hashing
In addition to single-cell analysis in whole and intact cells, single-nucleus analysis makes it possible to characterize cellular states and physiology in tissues that are challenging to dissociate. This includes tissues that are rich in certain cell types like neurons, adipocytes and muscle cells. Single-nucleus analysis can also help when tissue storage is required, as it is difficult to recover and obtain single-cell suspensions from archived frozen material.
To pool isolated nuclei from different samples, we developed nuclear hashing reagents. Our nuclear hashtag antibodies recognize a family of nuclear pore complex proteins and can cross-react with vertebrate organisms and other invertebrate species, such as Xenopus and yeast.
Read more about single nucleus hashing analysis in a Nature Communications paper first describing the technology by Gaublomme, et al.
Understand the Differences between Hashtag Formats
We offer hashtag reagents in our TotalSeq™-A, B, and –C formats. Hashtag reagents from all formats will function similarly, but the workflow and required primers differ.
For TotalSeq™-A reagents, the PCR handle between antibodies and hashtag reagents are different. In practice, this means that you will generate two separate libraries– one for your cell surface markers (referred to as ADTs) and one for hashtag-derived oligos (referred to as HTOs). This is beneficial because the two libraries can be mixed at different ratios prior to sequencing to avoid an excess of reads from the hashtags.
For TotalSeq™-B and –C reagents, the PCR handle is the same between both your antibodies of interest and the hashtag antibodies. In practice, this means that the hashtag and antibody libraries must be prepared together. To avoid an excess of HTO readings, we recommend titrating down the hashtag reagents. For more information on how to titrate TotalSeq™ products, read our FAQs.
Simplify Single-Cell Multiomic Workflows with TotalSeq™ Antibody Cocktails
Gain flexible cellular phenotyping with our TotalSeq™ antibody cocktails. These pre-defined cocktails integrate seamlessly with various single-cell RNA sequencing workflows, allowing you to analyze from nine to hundreds of proteins simultaneously, depending on your experimental goals. Each cocktail is expertly designed with pre-titrated amounts of lyophilized antibodies to optimize your workflow.
Benefits of using antibody cocktails:
-
Provided in convenient, single-use tubes.
-
Antibodies are pre-titrated and balanced using next-generation sequencing as the readout for optimal performance.
-
Reduces inconsistences associated with pipetting and handling multiple vials and samples.
-
Lower cost compared to buying individual antibodies.
-
Minimizes variability between different experiments, labs, and longitudinal studies.
Quick Selection Guide

TotalSeq™ Cocktail Descriptions
| TotalSeq™-A | TotalSeq™-B | TotalSeq™-C | ||
| Assay Compatibility | TotalSeq-A cocktails are compatible with single-cell platforms that use poly(dT) oligonucleotides as the mRNA capture method, and 10x Genomics Chromium Universal 3' Gene Expression assay. | TotalSeq- B cocktails are compatible with 10x Genomics Chromium Universal 3’ and Flex Gene Expression assays. | TotalSeq-C cocktails are compatible with 10x Genomics Chromium Universal 5’ and Flex Gene Expression assays. | |
|
Human |
Human Universal Cocktails V2.0 New |
TotalSeq™-A Human Universal Cocktail, V2.0 175 primary antibodies and 9 isotype controls. |
TotalSeq™-B Human Universal Cocktail, V2.0 Coming Soon |
TotalSeq™-C Human Universal Cocktail, V2.0 175 primary antibodies and 9 isotype controls. |
| Human Universal Cocktails V1.0 |
TotalSeq™-A Human Universal Cocktail, V1.0 154 primary antibodies and 9 isotype controls. |
TotalSeq™-B Human Universal Cocktail, V1.0 134 primary antibodies and 6 isotype controls. |
TotalSeq™-C Human Universal Cocktail, V1.0 130 primary antibodies and 7 isotype controls. |
|
|
Human Essential Cocktails V1.0 New |
TotalSeq™-A Human Essential Cocktail, V1.0 100 primary antibodies and 9 isotype controls. |
TotalSeq™-B Human Essential Cocktail, V1.0 Coming Soon |
TotalSeq™-C Human Essential Cocktail, V1.0 100 primary antibodies and 9 isotype controls. |
|
| TBNK Cocktails | TotalSeq™-B Human TBNK Cocktail | TotalSeq™-C Human TBNK Cocktail | ||
| 9 primary antibodies for the identification of principal lineage immune cell types. | ||||
| Download the full barcode list | Download the full barcode list | Download the full barcode list | ||
| Mouse
|
Mouse Universal Cocktails V1.0 |
TotalSeq™-A Mouse Universal Cocktail, V1.0 119 primary antibodies and 9 isotype controls. |
TotalSeq™-B Mouse Universal Cocktail, V1.0 95 primary antibodies and 7 isotype controls. |
TotalSeq™-C Mouse Universal Cocktail, V1.0 113 primary antibodies and 9 isotype controls. |
| Mouse Myeloid Cocktail V1.0 |
|
TotalSeq™-B Mouse Myeloid Cocktail, V1.0 76 primary antibodies and 8 isotype controls. |
||
Characterize Cell Diversity to an Unprecedented Level with TotalSeq™ Cocktails
Universal Cocktails
TotalSeq™ Human and Mouse Universal Cocktails range in size containing from 95 to 175 primary antibodies and their associated isotype controls for large-scale protein detection.
Version 2.0 of our TotalSeq Human Universal Cocktails includes up to 63 additional antibodies compared to Version 1.0. This update provides greater dimensionality and a more comprehensive view of cellular heterogeneity, function, and interactions within complex biological systems. The selection of antibodies was informed by high-impact peer-reviewed publications and feedback from the scientific community, aiming to broaden the coverage of surface markers pertinent to translational research and immuno-oncology applications.

Major immune cell lineages and selected cell states are identified using the TotalSeq™-A Human Universal Cocktail, V2.0. Human peripheral blood mononuclear cells (PBMCs) were stained with the TotalSeq™-A Human Universal Cocktail, V2.0, and processed using the 10x Genomics Single Cell 3’ v3.1 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Cell clusters were identified based on protein expression only. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions.
Essential Cocktails
The TotalSeq™ Essential Cocktails target 100 proteins, in contrast to the 175 proteins identified by the TotalSeq Human Universal Cocktails V2.0. They focus on the most frequently cited proteins for characterizing principal and sub-lineage immune cell types, while maintaining high multiplexing capacity. Additionally, they include proteins highly relevant to translational research and immuno-oncology.

Major immune cell lineages and selected cell states are identified using the TotalSeq™-C Human Essential Cocktail, V1.0. Human peripheral blood mononuclear cells (PBMCs) were stained with the TotalSeq™-C Human Essential Cocktail, V1.0 and processed using the 10x Genomics Single Cell 5’ v2 feature barcoding kit and Illumina sequencing. Protein and RNA count data were transformed and visualized in a UMAP projection for each component of the cocktail. Cell clusters were identified based on protein expression only. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions.
Mouse Myeloid Cocktail
The TotalSeq™-B Mouse Myeloid Cocktail has been optimized on enzymatically digested mouse splenocytes to aid in the multiomic characterization of myeloid lineage cell types, and ideal for samples depleted of lymphoid lineage cells.
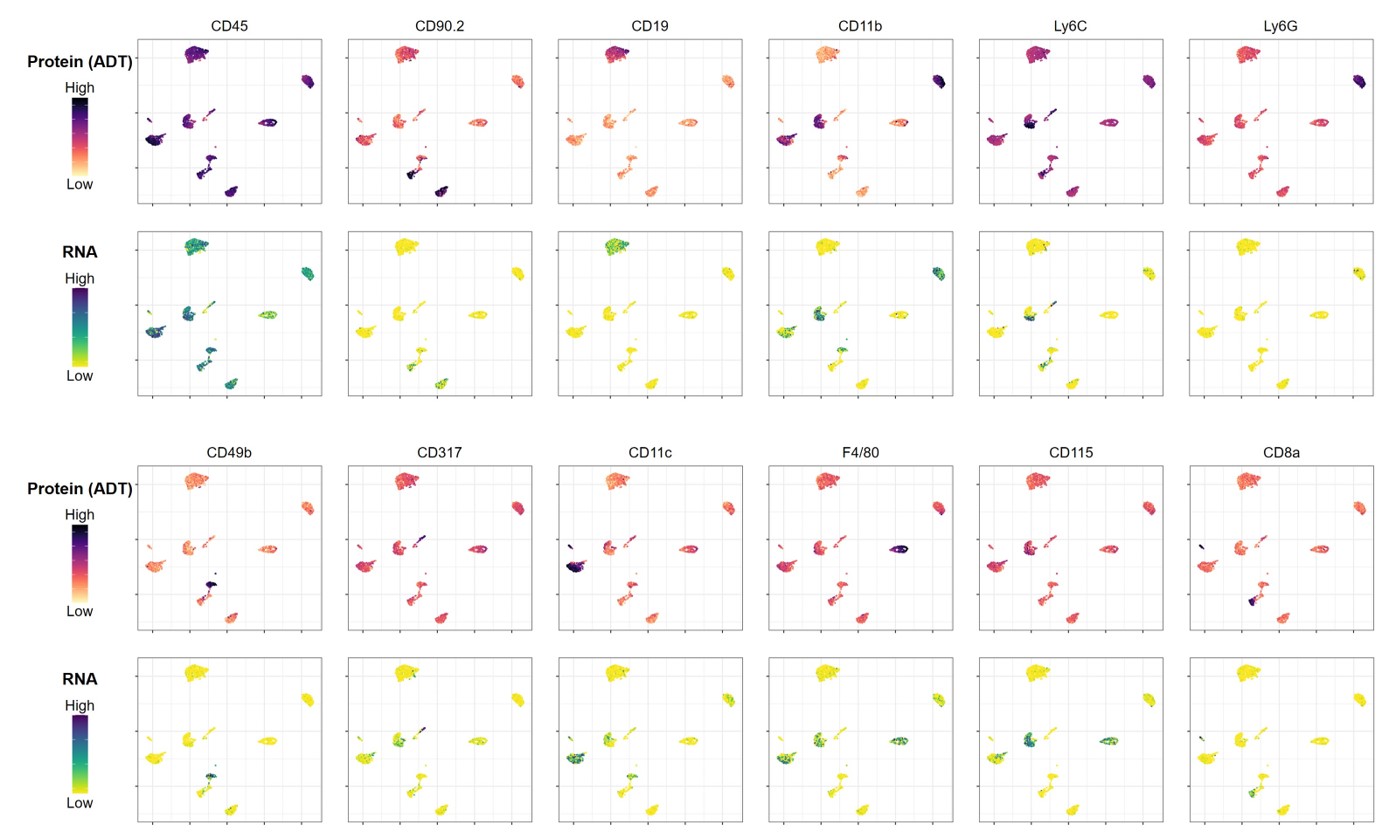
Data shown for a subset of markers in the cocktail; download the complete dataset for all 76 primary targets and 8 isotype controls.
Major immune cell lineages and selected cell states are identified using the TotalSeq™-B Mouse Myeloid Cocktail. Enzymatically dissociated mouse splenocytes were depleted of T and B cell populations, stained with the TotalSeq™-B Mouse Myeloid Cocktail and processed using the 10x Genomics Single Cell 3’ v3.1 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Cell clusters were identified based on protein expression only.
TBNK Cocktails
Our TBNK Panels are designed to react with 9 primary antibodies for the identification of T, B, and NK cells as defined by the expression of CD3, CD4, CD8, CD11c, CD14, CD16, CD19, CD45, and CD56.

Human PBMCs were stained with the TotalSeq™ Human TBNK panel containing antibodies against CD3, CD4, CD8, CD19, CD56, CD14, CD11c, CD16, and processed using the 10x Genomics' Single Cell 3’ v3 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Clusters were identified based on protein expression only.








Follow Us