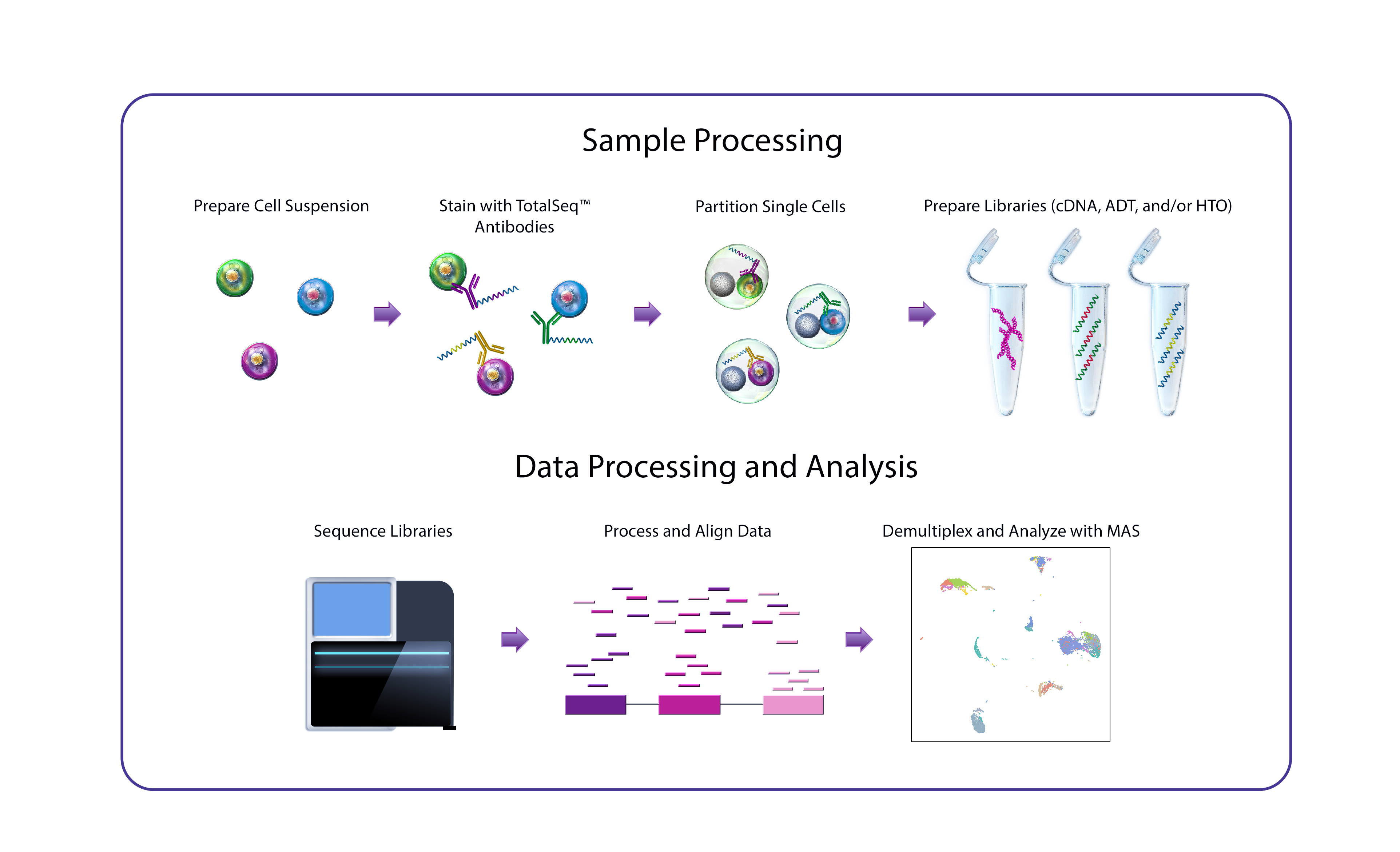
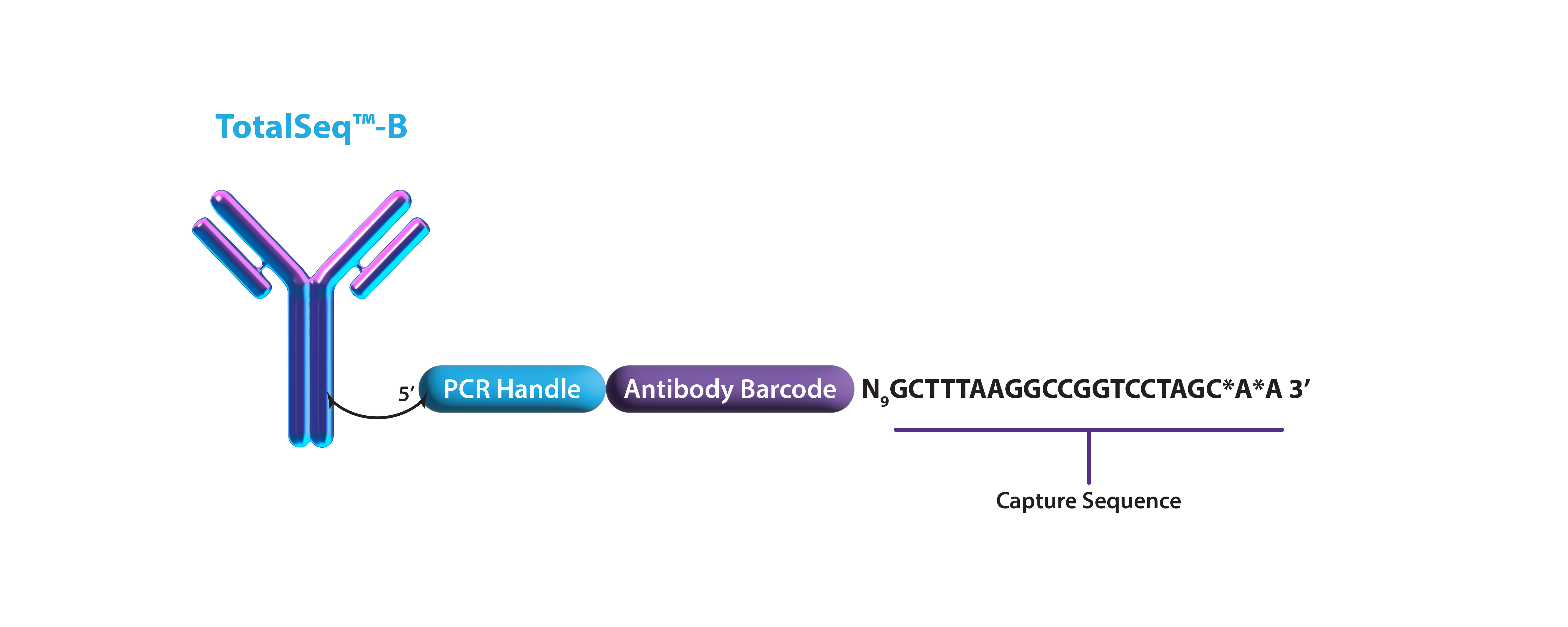
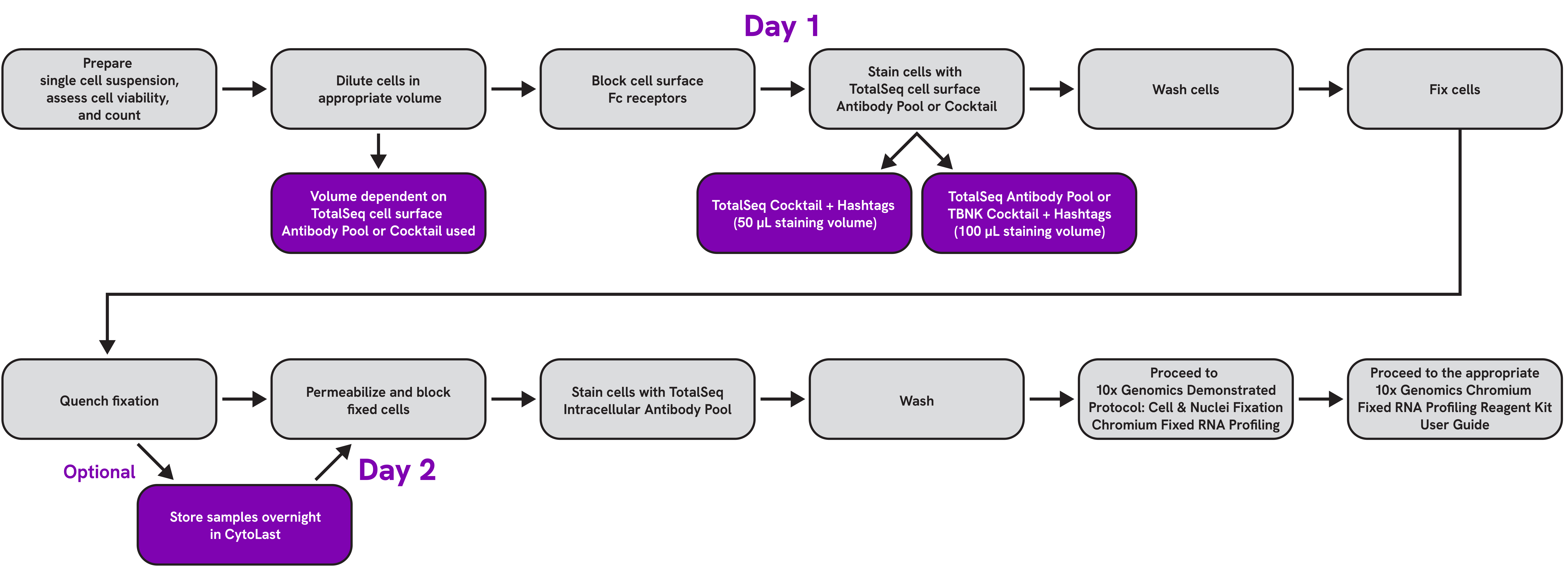
Characterize Cell Diversity to an Unprecedented Level with TotalSeq™ Cocktails
TotalSeq™ Human and Mouse Universal Cocktails range in size containing from 95 to 175 primary antibodies and their associated isotype controls for large-scale protein detection.
Version 2.0 of our TotalSeq Human Universal Cocktails includes up to 63 additional antibodies compared to Version 1.0. This update provides greater dimensionality and a more comprehensive view of cellular heterogeneity, function, and interactions within complex biological systems. The selection of antibodies was informed by high-impact peer-reviewed publications and feedback from the scientific community, aiming to broaden the coverage of surface markers pertinent to translational research and immuno-oncology applications.

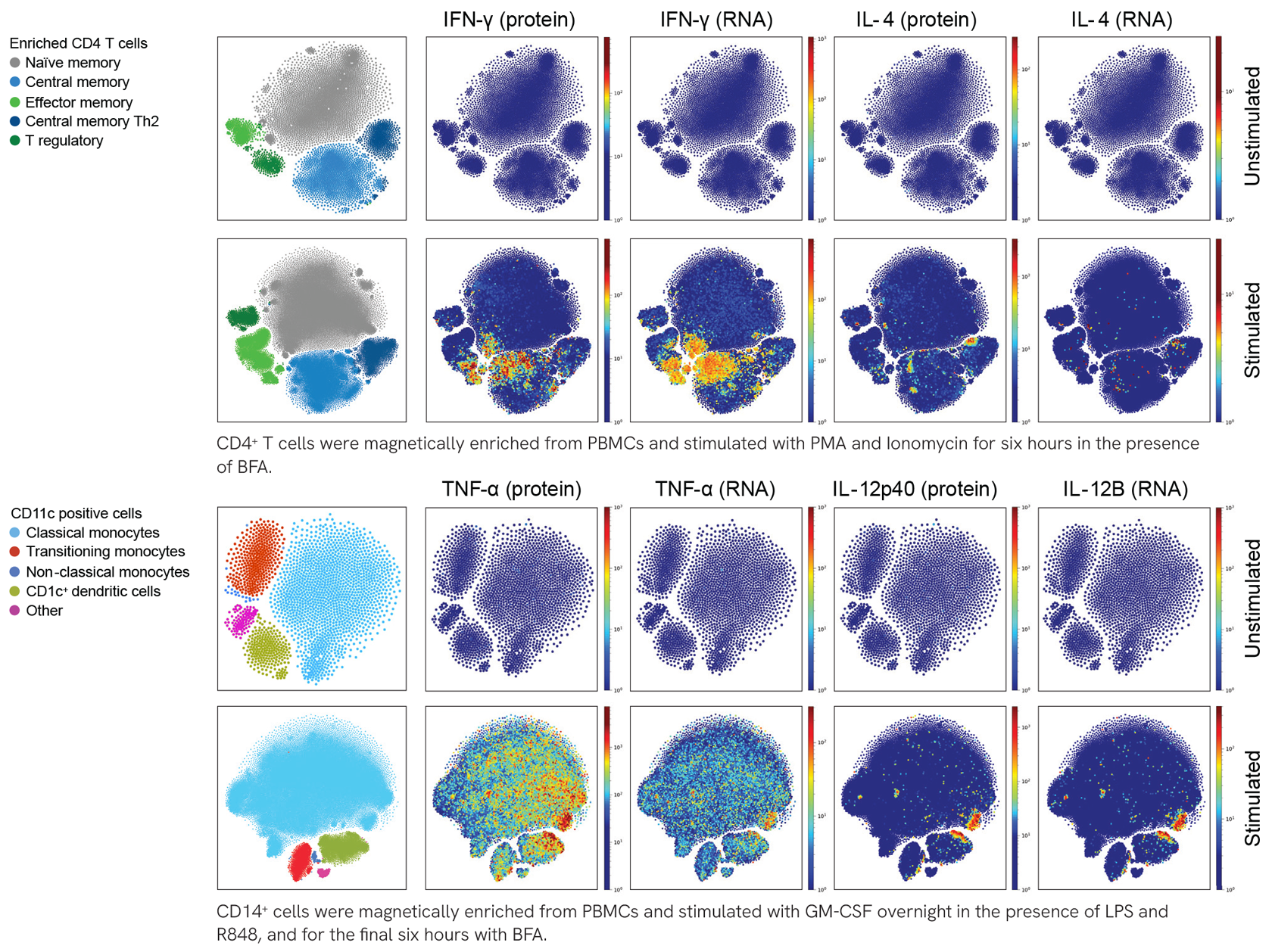
Major immune cell lineages and selected cell states are identified using the TotalSeq™-A Human Universal Cocktail, V2.0. Human peripheral blood mononuclear cells (PBMCs) were stained with the TotalSeq™-A Human Universal Cocktail, V2.0, and processed using the 10x Genomics Single Cell 3’ v3.1 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Cell clusters were identified based on protein expression only. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions.
The TotalSeq™ Essential Cocktails target 100 proteins, in contrast to the 175 proteins identified by the TotalSeq Human Universal Cocktails V2.0. They focus on the most frequently cited proteins for characterizing principal and sub-lineage immune cell types, while maintaining high multiplexing capacity. Additionally, they include proteins highly relevant to translational research and immuno-oncology.

Major immune cell lineages and selected cell states are identified using the TotalSeq™-C Human Essential Cocktail, V1.0. Human peripheral blood mononuclear cells (PBMCs) were stained with the TotalSeq™-C Human Essential Cocktail, V1.0 and processed using the 10x Genomics Single Cell 5’ v2 feature barcoding kit and Illumina sequencing. Protein and RNA count data were transformed and visualized in a UMAP projection for each component of the cocktail. Cell clusters were identified based on protein expression only. Refer to the complete dataset to see all cocktail components profiled on PBMCs under stimulation conditions.
The TotalSeq™-B Mouse Myeloid Cocktail has been optimized on enzymatically digested mouse splenocytes to aid in the multiomic characterization of myeloid lineage cell types, and ideal for samples depleted of lymphoid lineage cells.
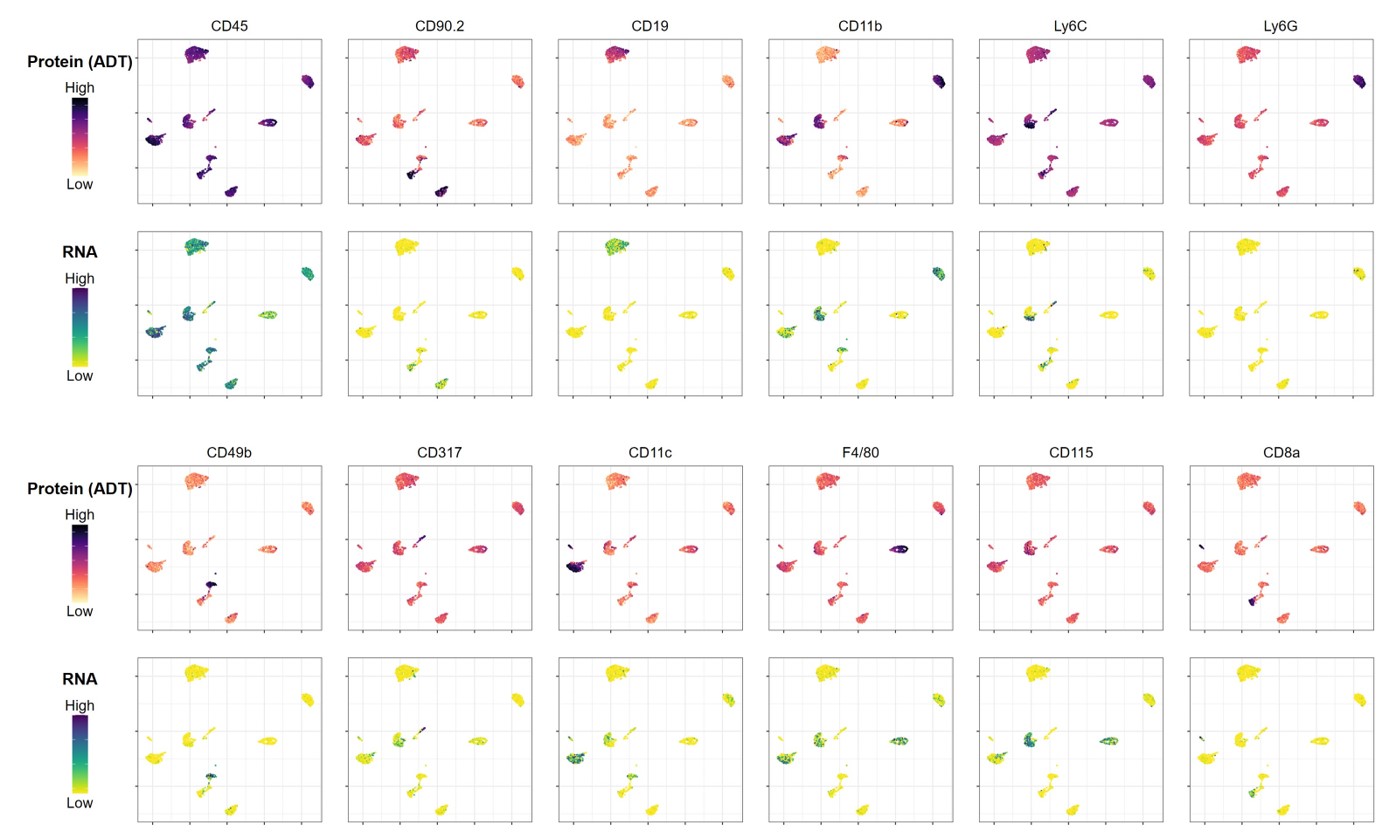
Data shown for a subset of markers in the cocktail; download the complete dataset for all 76 primary targets and 8 isotype controls.
Major immune cell lineages and selected cell states are identified using the TotalSeq™-B Mouse Myeloid Cocktail. Enzymatically dissociated mouse splenocytes were depleted of T and B cell populations, stained with the TotalSeq™-B Mouse Myeloid Cocktail and processed using the 10x Genomics Single Cell 3’ v3.1 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Cell clusters were identified based on protein expression only.
Our TBNK Panels are designed to react with 9 primary antibodies for the identification of T, B, and NK cells as defined by the expression of CD3, CD4, CD8, CD11c, CD14, CD16, CD19, CD45, and CD56.

Human PBMCs were stained with the TotalSeq™ Human TBNK panel containing antibodies against CD3, CD4, CD8, CD19, CD56, CD14, CD11c, CD16, and processed using the 10x Genomics' Single Cell 3’ v3 feature barcoding kit and Illumina sequencing. Protein count data were transformed and visualized in a UMAP projection overlaid with protein expression levels for each component of the cocktail. Clusters were identified based on protein expression only.
 Login/Register
Login/Register 














Follow Us